Review Article - Volume 2 - Issue 6
Role of molecular biology in cancer treatment
Ans Ahmad; Love Kumar; FNU Neelam; Marjan Assefi*
University of North Carolina at Greensboro, Joint School of NanoScience and Nano Engineering, North Carolina, USA.
Received Date : Oct 04, 2022
Accepted Date : Nov 14, 2022
Published Date: Dec 07, 2022
Copyright:© Marjan Assef 2022
*Corresponding Author : Marjan Assefi, University of North Carolina at Greensboro, Joint School of NanoScience and Nano Engineering, North Carolina, USA.
Email: massefi@aggies.ncat.edu
DOI: Doi.org/10.55920/2771-019X/1315
Abstract
Cancer is a genetic disease and essentially emerges because of various reasons incorporate enactment of onco-qualities, glitch of cancer silencer qualities or mutagenesis because of outer factors. Oncogenes are liberated type of typical proto-oncogenes expected for cell division, separation and guideline. The transformation of proto-oncogene to oncogene is caused because of movement, revamp of chromosomes or change in quality because of expansion, cancellation, duplication or viral disease. These oncogenes are focused on by drugs or RNAi framework to forestall expansion of dangerous cells. There have been created various strategies of sub-atomic science used to analyze and treat disease, including retroviral treatment, hushing of oncogenes and changes in growth silencer qualities.
Keywords: Molecular Biology; RNAi; Viral; Oncogene; Chromosome; cancer.
Introduction
Cancer is a genetic disease. The statement of oncogenes isbeing a significant occasion in beginning phases of cancer development. Oncogenes are actuated through two instruments: either by contamination of cells by growth infections or by transformation of cell proto-oncogenes (which are generally typical) to oncogenes. Then cancers start by oncogenic change of just a solitary cell. A cancers embrace the capacity to get away from the site of their starting point and encroach different pieces of the body. This interaction is called metastasis. Strong cancers for example sarcomas, could be moved starting with one creature then onto the next utilizing rous sarcoma infection. Cancers could be caused either by the expansion or by articulation of hereditary material, which for this situation was viral DNA, to typical cells. Rous was introduced a nobel prize for his work. In 1978, growths of nonverbal beginning were likewise found [1].
Discussion
Oncogenes in cancer development
Oncogenes are the growth causing qualities and play significant part being developed of numerous tumors. In 1970, SRC oncogene was found in chicken retrovirus. Because of some change in the generally typical proto-oncogenes, their liberation happens and uncontrolled multiplication of cells starts and prompts malignant growth [4]. At genomic level, just single oncogenic allele is expected to adjust typical quality capability on account of its prevailing property. The beginning of oncogene can be cell i.e., from inside the body or viral i.e., from some infection [3].
Gene duplication, expansion, inclusion, cancellation or chromosomal movement, chromosomal revision of specific proto-oncogenes adjusts their capability and converts them into oncogenes. These changes overexpress the protein to an uncontrolled level, which might prompt cancer. These changes might happen because of outside factors or inner elements or both like viral contamination, radiation or synthetic compounds, injury and illness [4]. Among these changes, viral disease is the uncommon reason for oncogene enactment in creatures however is vital for understanding oncogene capability.
Viral infection
Retroviruses or DNA infections cause viral contaminations. These infections taint the host either by embedding oncogenes in have chromosome, meddling proto-oncogene record factors/controllers or by embedding homologous successions relating to typical proto-oncogene of host. For instance, retrovirus conveying SRC oncogenes contaminates the host, coordinates viral chromosome in have chromosome, further partitions the viral descendants and taints the encompassing cells, actuating overexpression of cell typical qualities and liberated expansion of cells to cause malignant growth [5].
Types and classification of oncogenes
Oncogenes can be arranged into five classes in view of protein items framed by transformation or liberation of proto-oncogenes. These incorporate development factors, development chemical/factor receptors (GRFs), serine/threonine kinases, ATPase atoms and, record factors. Transformations in the development elements can prompt a few kinds of malignant growths, for example, fibro sarcoma, glioblastoma (cerebrum disease), osteosarcoma (bone disease) and so on [6,7].
In a few cancers, "ligand-restricting space" erasures of Epidermal Development Component Receptor cause progressive enactment of receptor even without ligand by transmembrane protein conveying tyrosine-kinase movement. This enactment causes communication with additional cytoplasmic proteins like "SRC area" and prompts liberation of various flagging pathways. For the most part in gastrointestinal, bosom and cellular breakdowns in the lungs, GFR changes happen [4]. Additionally, overexpression of Raf-1 kinase and cyclin-subordinate kinases because of uncontrolled phosphorylation might cause numerous tumors like thyroid and ovarian disease.
Liberated initiation of Gases, for example, Raps, causes actuation of MAPK pathway and uncontrolled flagging and division of cells cause a few tumors like myeloid leukemia. Record factor proteins are results of proto-oncogenes. The change, movement or modification of these causes overexpression of quality and undesirable successive record of target quality that prompts any kinds of tumors, for example, pancreatic and cellular breakdown in the lungs [6].
Role of oncogenes to treat cancer
The oncogenes are focused on to treat oncogenic malignant growth. A few oncogenes examined above are designated by medications and quality treatments to repress, capture, manage or senescence their qualities. For instance, Imagine (ABL kinase inhibitor) or Greeves is utilized to treat BCR-ABL. Gefitinib or Iressa, erlotinib or Tarceva are utilized to target EGFR. VEGF oncogenes are designated by bevacizumab or sorafenib. Sorafenib is additionally used to downregulate or repress B-Raf oncogene. These specialists/drugs are utilized, at times in mix, for chemotherapy to hinder expansion of oncogenes or to downregulate flagging oncoproteins in a few flagging pathways to treat oncogenic malignant growths [8]. Notwithstanding, it is hard to target "non-kinase oncogenes" through medications like Myc and Ras.
Tumor suppressor genes in cancer
Tumor suppressors assume their part by restraining cell expansion and cancer improvement. In the greater part of the growths, inactivation of the growth silencer qualities kills the negative guideline of these qualities over cell multiplication that prompts strange cell expansion, accordingly, causing disease. Growth silencer qualities have "loss of capability" transformations since they foster disease by inactivating their inhibitory impact on cell multiplication. For a growth silencer quality to advance cancer improvement, the two duplicates of the quality should be inactivated on the grounds that one duplicate is adequate for controlling cell expansion. These changes act passively [9].
Role of Tumor Suppressor Genes in Cancer Wilms’ Tumor 1 Gene
Some tumor-suppressing genes go about as transcriptional administrative proteins. For instance, the result of WT1 quality which is a repressor protein and acts by smothering record of numerous development factor-inducible qualities. WT1 is made idle in Wilms' growths (which is a cancer in kidney tracked down in youngsters). Insulin-like development factor II is the objective of WT1 quality, over-communicated in Wilms' growths, in this way adding to unusual cell multiplication [11].
Retinoblastoma and INK4 Genes
Several tumor suppressor genes regulate cell cycle progression through a specific stage e.g. manage cell cycle movement through a particular stage for example protein results of Rb and INK-4 qualities. Retinoblastoma is the growth of the eye. Two mutagenic occasions are expected for the retinoblastoma improvement in irregular cases while just a single mutagenic occasion is required in people with acquired type of the sickness in which it shows autosomal prevailing legacy. In typical cells, Cdk2 and cyclin D edifices control the passage through the limitation point, in this way phosphorylating and inactivating pRb. pRb additionally blocks the passage through the limitation point in the G1 period of the cell cycle by quelling the record of numerous qualities engaged with cell cycle progression. The INK4 cancer silencer quality likewise controls development through the limitation point by encoding Cdk inhibitor p16. Inactivation of INK4 brings about uncontrolled phosphorylation of Rb [10].
p53 Tumor Suppressor Gene
The p53 assumes its part by managing cell cycle and modified cell passing. The p53 can capture the cell cycle upon DNA harm. It permits the DNA to fix or cause the customized cell passing (apoptosis). This is accomplished by enacting various qualities associated with controlling and managing the cell cycle. Transformation in p53 in tumorigenic cells brings about uncontrolled cell multiplication and wasteful DNA fix. p53 changes are assessed to be the most widely recognized in cancers of people, roughly half or much more noteworthy than that [12].
Breast Cancer-1 and 2 Genes
Breast cancer-1 and 2 genes are connected to familial bosom disease. Bosom Malignant growth 1 quality comprises of 100 Kb DNA and 21 exons. It has a zinc-finger space like that in the DNA restricting proteins. Bosom malignant growth 1 is a cancer silencer quality. BRCA-2 is situated on chromosome 13.
Tumor Suppressor Genes and their application
Tumor suppressor genes can be learned at the degrees of DNA, mRNA, and proteins in the ordinary and destructive cells utilizing different strategies. Tests for the identification of heterozygosity can be useful for recognizing people inclined toward retinoblastoma and different malignancies. Higher recurrence of p53 transformations additionally offers indicative and insightful conceivable outcomes. PCR enhancement can be utilized to concentrate on the progressions alongside late strategies, for example, RNase insurance tests, single-strand conformational polymorphism or denaturing gel electrophoresis. Immunometric-type examines are very great at estimating changed p53 in cancer cell line lysates and tissue homogenates [13,14].
Molecular pathology: Diagnosis of cancer
One of the essential difficulties in the clinical administration of malignant growth patients is to lay out the right conclusion. Therefore, various advancements have been created and are currently regularly utilized to subtype atomically malignant growths. These incorporate immunohistochemistry, immunofluorescence, and the examination of DNA and RNA removed from the sore through In situ hybridization and fluorescent in situ hybridization (FISH). The disease example is then subtyped utilizing various methodologies of atomic science including Sanger sequencing, pyrosequencing, allele-explicit PCR. Malignant growth genotyping is performed by depiction test, mass spectroscopy based measures and cutting edge sequencing (in light of fluorescence or semiconductor locators). The presentation of cutting edge sequencing is effectively revealing the genuine variety of malignant growths as well as to characterize repeating changes focused on with new treatments. Such genomic-level examinations will keep on having an effect for a long time [15,16].
Cancer treatment – then & now
Different therapy methods and treatments have been applied for the therapy of disease at various times. Probably the most well-known strategies utilized incorporate a medical procedure, radiation treatment, chemotherapy, hormonal treatment, immunotherapy, adjuvant treatment, designated development signal restraint, sedates that initiate apoptosis, nanotechnology, RNA articulation and profiling, and the most recent being CRISPR [17]. Not many of these will be subsequently examined in this survey. Malignant growth cells can likewise be killed by quality substitutions or by taking out oncogenes. Oncolytic infections can be utilized in blend with chemo-remedial specialists to annihilate disease cells also [18].
Retroviral therapy for cancer
Aside from the traditional strategies, retroviruses (RVs) have additionally been utilized in disease treatment. RVs can be and have been utilized for moving qualities to mammalian cells. Most famous RVs are the ones gotten from the Moloney Murine leukemia infection (MoMLV). Over the most recent twenty years, counterfeit advancement of RVs has empowered their applications in creating transgenic creatures, stable conveyance of siRNA and clinical preliminaries for quality treatment. The extraordinary capability of RVs was examined in ongoing reports about the effective clinical preliminaries of quality treatment in patients with extreme immunodeficiency sickness. Nonetheless, there are a few plausible dangers intrinsically related with that [19]. A similar examination was finished by planning two gatherings of vectors. One was naturally replicative, the other was deficient thus it had a partner retrovirus with it. Under in vitro conditions, the replicative infections accomplished over 85% transduction while the other transduced just under 1%. This analysis plainly shows the capability of RRVs for creating malignant growth quality treatment [20].
Retroviral tagging and insertional oncogenesis
As of late, premium in exploring retroviral vector additions (insertional oncogenesis) has been developing. For a long time, viral inclusion destinations were utilized to distinguish potential oncogenes and malignant growth flagging pathways. The extent of this approach has been widened by strategies like inclusion site cloning by high throughput PCR, accessibility of hereditarily adjusted creatures and with the fulfillment of mouse genome project [21,22]. Be that as it may, various specialists have distinguished many normal destinations. These reconciliation locales are generally connected with disease qualities in MoMLV-prompted murine haematopoietic malignancies [23-25]. For the most part, larger part of the additions exist outside the coding districts. Subsequently, just under 10% of the RISs can be considered as the acknowledged growth silencer qualities. Strangely, around 17%-18% of RISs are designated record factors [19].
Problems with Retroviral Therapy
There are some wellbeing concerns related with the retroviral quality treatment tended to. A few potential answers for this issue could be designated disease, transcriptional focusing on, neighborhood conveyance, co-transduction of retroviral vector with a self-destructive quality, explicitly focused on retroviral inclusion, and SIN vectors. These methods, with both viral and non-viral frameworks, can be utilized in conventions for quality treatment. In any case, insertional oncogenesis stays the main issue with respect to retroviral quality treatment [26]. A protected and quick method for designated approach could be the utilization of frothy infections [27,28]. These infections are innocuous to people but have many hosts [29-31]. Notwithstanding, for ex vivo cell-based quality treatment, insertional oncogenesis can be stayed away from by pre-screening of transduced cells to choose just those cells (clones) which have the transgene just at a positive site of the chromosome [26].
Molecular biology techniques for the treatment of cancer
At first, homologous recombination was utilized to inactivate the objective quality. This was finished for describing the capability of qualities. This technique was not however useful as it seemed to be not effective in that frame of mind onto the objective site. It was extremely extensive cycle, the choice interaction was relentless and there were numerous serious mutagenic impacts of this strategy [32].
RNA Interference
One of the techniques utilized in malignant growth treatment is RNA impedance. RNA impedance includes the utilization of little non-coding RNA that can tie with different mRNAs and can hinder their interpretation into proteins. This can bring about the deficiency of capability of qualities. In malignant growth treatment, these RNAi can be utilized to annihilate the capability of disease qualities that keep malignant growth from spreading [33-35]. The RNAi technique is quick, modest and it has a high productivity so it supplanted homologous recombination strategy. In any case, it has disadvantages that incorporate deficient knockdown and impermanent avoidance of quality capability. It likewise radiates target impacts unpredicted. Malignant growth repeat was likewise found now and again. This multitude of issues lead the analysts to investigate new strategies to change the quality capability [36].
Genome altering is a far superior and new strategy to treat disease. Researchers utilized designed nucleases that have explicit spaces that can tie to the objective site followed by its cleavage [37, 38]. These nucleases had the option to actuate twofold strand breaks (DSBs) in the objective followed by the enactment of DNA fix systems. Two sorts of nucleases are utilized that incorporate programmable nucleases like Zinc Finger Nucleases (ZFNs) and record activator-like effector nucleases (TALENs). These nucleases were fruitful in genome altering for relieving disease in various creature models [39].
Zinc finger proteins (ZNFs)
ZNFs are the principal nucleases to be utilized for quality altering
These are found as DNA restricting spaces in eukaryotes. These are comprised of 30 amino acids modules organized as a variety of Cys2-His2 DNA-restricting zinc fingers. These modules are utilized to a nuclease space of FokI [37,40,41]. The modules comprise of 3-6 zinc fingers that can distinguish nucleotide trios [42,43]. The FokI nuclease works just as dimers so a couple of zinc finger nucleases is expected to focus on any district in the genome. One ZFN will recognize the arrangement upstream of the genome district to be changed and other will distinguish downstream grouping [44]. These exhibits tie to local DNA successions that are in the contrary strands to prompt a twofold abandoned break in the particular district. The breaks are then fixed by various techniques that can cause various changes in the particular district like point transformations, indels or movements. The ZNFs are specially crafted with the goal that they perceive every single imaginable nucleotide and a particular locale of DNA [42,45,46].
Transcription activator-like effector nucleases (TALENs)
TALENs are like zinc finger nucleases, as they additionally need DNA restricting themes and a similar nuclease to alter the genome. The distinction lies in the acknowledgment of nucleotides as the TALENs space distinguishes just a single nucleotide rather than a trio. The connections between the TALEN spaces and their objective locales are more grounded when contrasted with ZNFs. It is simpler to plan TALENs when contrasted with planning of ZNFs [46,47]. Involving TALENs for malignant growth treatment is extremely successful strategy. It needs two explicitly designed TALENs that can distinguish the arrangements of DNA in the objective quality on the contrary strands. It dimerizes the FokI nuclease cleavage space in the TALENs, separating the arrangement in the objective quality [48-50]. This causes twofold abandoned DNA breaks in the designated quality. The injury because of the DNA break is fixed toward the end-joining DNA fix framework. The objective quality is modified because of the adjustment of the understanding edge. This technique can likewise be utilized to eliminate the all around present transformations. This quality altering innovation can be utilized to treat the disease cell lines effectively as it can focus on any quality in the genome. Complex malignant growth qualities can be treated by utilizing TALENs [51].
CRISPR/CAS9 system: A powerful tool for genome editing in cancer
A strong genome-altering innovation known as Bunched routinely interspaced palindromic successions abbreviation CRISPR, is presently obscuring any remaining genome-designing methods. This progressive method permits analysts to achieve designated control in any quality (DNA grouping) in the whole genome of any life form in vitro or presently even straightforwardly in endogenous genome, accordingly assisting with explaining the utilitarian association of genome at frameworks level and distinguishing relaxed hereditary varieties. CRISPR assumes a fundamental part in discovery of tumors [52].
Mechanism of CRISPR-Cas9 in cancer treatment
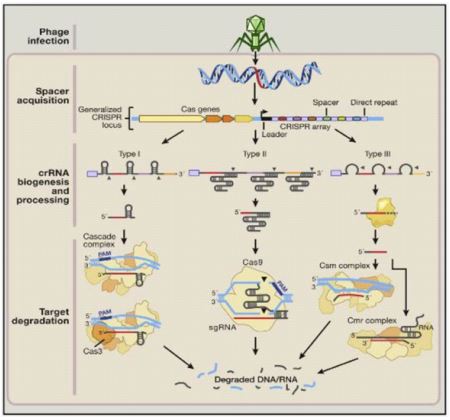
In the event that malignant growth causing quality is known, dangerous cells can be treated with CRISPR-Cas9 framework which assists in quality erasure and supplanting with a typical quality. Yin et al in his paper talk about the infusion cycle utilizing CRISPR-Cas9 framework to cut and bring quality into liver cells. CRISPRCas9 is more powerful for single quality transformation tumors and is for the most part conveyed in vitro in a specific area. In the event of metastatic tumors, in vitro conveyance becomes troublesome. CRISPR principally comprises two organic parts: Designed single aide RNA (sgRNA) and Cas9. A little aide RNA (crRNA and tracrRNA) is utilized to perceive the correlative grouping explicit objective flanked by proto-spacer nearby theme (PAM) and it guides endonucleases for example Cas9 to divide this arrangement [53]. Different CRISPR-Cas frameworks have been gathered significantly into three kinds (I-III) and subtypes (as I-E) contingent upon assorted bacterial and archeal rehash groupings, as qualities, and their method of activity [54]. Type I and III frameworks share a typical system of handling of pre-crRNAs (to crRNA) by means of specific Cas endonucleases, and on development, each crRNA complex with multi-Cas protein is fit for perceiving and separating objective successions which are reciprocal to crRNA. In as opposed to this, Type II framework is viewed as the core of genome designing device since it includes decreased number of Cas chemicals.
It requires non-coding tracrRNA which sets off the handling of pre-crRNA through dsRNA explicit Ribonuclease RNase III and cas9 protein (just protein doing viable crRNA-intervened hushing of target) [55]. CRISPR loci are dominatingly made out of different rehashed successions (approx. 21-48 bp) interspaced by factor spacer successions (approx. of 2-72 bp) and Cas qualities arranged close by CRISPR locus. In the first place, this CRISPR locus exhibit is deciphered as single RNA, handling of this delivered pre-RNA (from inside recurrent groupings) into solitary more limited units of CRISPR RNAs (crRNAs) utilizing host proteins is finished. Mature crRNA successfully ties to nucleases for example Cas proteins, consequently this complex of crRNA-cas9 helps in perceiving and afterward dividing the objective attacking DNA or RNA having complementarity to crRNA (destinations called protospacers). A theme of 2-5 nucleotide called as PAM is situated close by protospacer in CRISPR-Cas frameworks I and II [54]. PAM, a nucleic corrosive grouping comprised of NGG or Bother trinucleotide for Cas9, flanks at 3' finish of the DNA target site, helps Cas9 in its particular cleavage action and furthermore works with Cas9 in distinctive self-versus non self-bacterial successions as PAM [52]. SpCas9 (called as Streptococcus pyogenes Cas9) is being used presently for viable genome altering in eukaryotic organic entities including people. PAM found downstream of target is just arrangement permitting cas9 target site determination. Cas9 proteins shift in their size and succession and have normal areas as HNH and RuvC endonuclease to separate two strands of target DNA Cas9HNH space is determined for cleavage of strand correlative to direct (target) grouping while Cas9 RuvC areas for non-corresponding (non-target) strand, likewise, Cas9 holds some saved arginine-rich destinations for restricting of nucleic corrosive [53]. Target acknowledgment by this crRNA coordinates the hushing of unfamiliar groupings through case proteins that capability in complex with crRNAs [52,54].
The Streptococcus pyogenes-determined CRISPR-Cas9 RNA-directed DNA endonuclease is confined to a particular DNA succession through a solitary aide RNA (sgRNA) grouping, which base matches with a particular objective grouping that is contiguous a protos-pacer nearby theme (PAM) grouping as NGG or Bother.
On enlistment of twofold abandoned breaks or scratches at designated districts, fixing is finished by either Non-homologous end joining (NHEJ) or Homology-coordinated fix (HDR) pathway. NHEJ is a mistake inclined fix system where joining of broken closes happens, which by and large outcomes in heterogeneous indels (additions and erasures) though HDR is an exact fix strategy wherein homologous giver format DNA is being utilized in fix DNA harm target site [53].
Advantages of CRISPR over traditional methods
The CRISPR/Cas9 framework is liked over the ZNFs and TALENs in view of many benefits. First and foremost, the objective plan process is less difficult for CRISPR as it relies on the ribonucleotide complex development rather than DNA acknowledgment. It tends to be planned effectively and it is a lot less expensive than planning nucleases as this doesn't require various proteins for each objective and wipes out difficult cloning steps. This can be utilized to focus on a particular grouping in the genome. The CRISPR framework is substantially more proficient than ZFNs and TALENs. The RNA encoding Cas protein can be infused straightforwardly for adjusting the host genome. It isn't the case extended and difficult interaction when contrasted with customary techniques [12,51]. By utilizing CRISPR, we can present multiplexed changes. Numerous qualities can be transformed all the while by infusing with numerous gRNAs. This cycle is quicker when contrasted with different techniques. It doesn't acquaint awareness with DNA methylation so it very well may be utilized assuming the objective site is GC rich.
Conclusion
Molecular biology has quickly advanced somewhat recently than it has ever previously. Different malignant growth treatment strategies are arising and succeeding and with the improvement of ZNFs, TALENs, and CRISPR, researchers can focus on any grouping in the genome, even various qualities. This will give huge assist in the therapy of illnesses with loving malignant growth, keeping away from the dangers brought about by the past techniques.
References
- Gorge F. The Molecular Basis of Cancer. Bridgewater Rev. 1998; 17(2): 3-6.
- McCann J, Choi E, Yamasaki E, Ames BN. Detection of carcinogens as mutagens in the Salmonella/micro some test: assay of 300 chemicals. Pros Natl AccadSic USA. 1975; 72(12): 5135-9.
- Perry AR. Oncogenes. Van No Strand’s Scientific Encyclopedia. ells. 2001.
- Croce CM. Oncogenes and cancer. N Engle J Med. 2008; 358: 502-11.
- Perrott MA, Suzy G, Croce CM. Mechanisms of oncogene activation. Kure DW, Pollock RE, Weichsel Baum RR, et al. Holland-Frei Cancer Med, 6th. 2003.
- Sepik M. Oncogenic viruses and mechanisms of oncogenes is. Turk J Vet AminSic. 2012; 36(4): 323-9.
- Jived S, Ali M, Ali F, Anwar SS, Wajid N. Status of oxidative stress in breast cancer patients in Pakistani population. Adv Life Sci. 2015; 2(3): 115-8.
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and nononcogene addiction. Cell. 2019; 136(5): 823-37.
- Diamandis EP. Oncogenes and tumor suppressor genes: new biochemical tests. Crit Rev Clin Lab Sci. 1992; 29(3-4): 269-305.
- Park BH, Vogelstein B. Tumor-suppressor genes. Cancer Med. 2003; 6: 87-102.
- Englert C, Hou X, Maheswaran S, et al. WT1 suppresses synthesis of the epidermal growth factor receptor and induces apoptosis. EMBO J. 1995; 14(19): 4662-75.
- Soussi T. The p53 tumor suppressor gene: from molecular biology to clinical investigation. Ann N Y Acad Sci. 2000; 910: 121-37.
- Diamandis EP. Clinical applications of tumor suppressor genes and oncogenes in cancer. Clin Chim Acta. 1997; 257(2): 157-80.
- Ahmed T, Ahmed RS, Basharat MU, et al. Comparative study to access coagulation abnormalities in breast cancer. Adv Life Sci. 2013; 1(2): 96-103.
- Lopez-Chavez A, Thomas A, Rajan A, et al. Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology phase II basket trial. J Clin Oncol. 2015; 33(9): 1000-7.
- Khan MT, Afzal S, Rehman AU, Zeb T. Interleukin 10 (IL-10) promoter-1082 A> G polymorphism and risk of cancer: Meta-analysis. Adv Life Sci. 2015; 2(2): 67-73.
- Sudhakar A. History of cancer, ancient and modern treatment methods. J Cancer Sci Ther.2009; 1(2): 1-4.
- Ottolino-Perry K, Diallo JS, Lichty BD, Bell JC, McCart JA. Intelligent design: combination therapy with oncolytic viruses. Mol Ther. 2010; 18(2): 251-63.
- Barquinero J, Eixarch H, Perez-Melgosa M. Retroviral vectors: new applications for an old tool. Gene Ther. 2004; 11(1): S3-9.
- Solly SK, Trajcevski S, Frisén C, et al. Replicative retroviral vectors for cancer gene therapy. Cancer Gene Ther. 2003; 10(1): 30-9.
- Mikkers H, Berns A. Retroviral insertional mutagenesis: tagging cancer pathways. Adv Cancer Res. 2003; 88: 53-99.
- Dudley JP. Tag, you’re hit: retroviral insertions identify genes involved in cancer. Trends Mol Med. 2003; 9(2): 43-5.
- Lund AH, Turner G, Trubetskoy A, et al. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat Genet. 2002; 32(1): 160-5.
- Mikkers H, Allen J, Knipscheer P, et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002; 32(1): 153-9.
- Suzuki T, Shen H, Akagi K, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002; 32(1): 166-74.
- Yi Y, Jong Noh M, Hee Lee K. Current advances in retroviral gene therapy. Curr Gene Ther. 2011; 11(3): 218-28.
- Mergia A, Chari S, Kolson DL, et al. The efficiency of simian foamy virus vector type-1 (SFV-1) in nondividing cells and in human PBLs. Virology. 2001; 280(2): 243-52.
- Rethwilm A. Foamy virus vectors: an awaited alternative to gammaretro-and lentiviral vectors. Curr Gene Ther. 2007; 7(4): 261-71.
- Callahan ME, Switzer WM, Matthews AL, et al. Persistent Zoonotic Infection of a Human with Simian Foamy Virus in the Absence of an Intact orf-2Accessory Gene. J Virol. 1999; 73(11): 9619-24.
- Heneine W, Switzer WM, Sandstrom P, et al. Identification of a human population infected with simian foamy viruses. Nat Med. 1998; 4(4): 403-7.
- Schweizer M, Falcone V, Gänge J, Turek R, Neumann-Haefelin D. Simian foamy virus isolated from an accidentally infected human individual. J Virol. 1997; 71(6): 4821-4.
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005; 6(6): 507-12.
- Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002; 108(2): 135-44.
- Flintoft L. Animal models: Mastering RNAi in mice. Nat Rev Genet. 2011; 12(6): 380.
- Dow LE, Lowe SW. Life in the fast lane: mammalian disease models in the genomics era. Cell. 2012; 148(6): 1099-109.
- McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002; 3(10): 737-47.
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010; 11(9): 636-46.
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011; 188(4): 773-82.
- Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006; 40: 363-83.
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996; 93(3): 1156-60.
- Bibikova M, Carroll D, Segal DJ, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001; 21(1): 289-97.
- Maeder ML, Thibodeau-Beganny S, Osiak A, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008; 31(2): 294-301.
- Ramirez CL, Foley JE, Wright DA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008; 5(5): 374-5.
- Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci USA. 1998; 95(18): 10570-5.
- Bhakta MS, Henry IM, Ousterout DG, et al. Highly active zinc-finger nucleases by extended modular assembly. Genome Res. 2013; 23(3): 530-8.
- Sander JD, Dahlborg EJ, Goodwin MJ, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods. 2011; 8(1): 67-9.
- Campbell JM, Hartjes KA, Nelson TJ, Xu X, Ekker SC. New and TALENted genome engineering toolbox. Circ Res. 2013;113(5): 571-87.
- Santiago Y, Chan E, Liu P-Q, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105(15): 5809-14.
- Bedell VM, Wang Y, Campbell JM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012; 491(7422): 114-8.
- Boch J. TALEs of genome targeting. Nat Biotechnol. 2011; 29(2): 135-6.
- Wu X, Blackburn P, Tschumper R, Ekker SC, Jelinek DF. TALEN-mediated genetic tailoring as a tool to analyze the function of acquired mutations in multiple myeloma cells. Blood Cancer J. 2014; 4(5): e210.
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPRCas9 for genome engineering. Cell. 2014; 157(6): 1262-78.
- Mao XY, Dai JX, Zhou HH, Liu ZQ, Jin WL. Brain tumor modeling using the CRISPR/Cas9 system: state of the art and view to the future. Oncotarget. 2016; 7(22): 33461-71.
- Bondy-Denomy J, Davidson AR. To acquire or resist: the complex biological effects of CRISPR–Cas systems. Trends Microbiol. 2014; 22(4): 218-25.
- Way JC, Collins JJ, Keasling JD, Silver PA. Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell. 2014; 157(1): 151-61.

