Research Article - Volume 3 - Issue 5
Synthesis and anticancer activities of Cu2+, Zn2+, Co2+ and Ni2+ complexes containing pyridine carboxylic acids
Li Zhang; Mei Luo*
Department of Chemistry and Chemical Engineering, Hefei University of Technology, Hefei, 23000, P.R. China
Received Date : Sep 12, 2023
Accepted Date : Sep 28, 2023
Published Date: oct 09, 2023
Copyright: © Mei Luo 2023
*Corresponding Author : Mei Luo, Department of Chemistry and Chemical Engineering, Hefei University of Technology, Hefei, 23000, P.R. China.
Email: luomei@pku.edu.cn
DOI: Doi.org/10.55920/2771-019X/1562
Abstract
Six metal complexes [CuC20H20N6O10] (Ⅰ), [ZnC7H9NO7] (Ⅱ), [CoC21H18N3O15] (Ⅲ), [CoC14H10N2O10] (Ⅳ), [NiC14H10N2O10] (Ⅴ) and [ZnC14H10N2O10] (Ⅵ) have been synthesized under one-pot self-assembly condition with 2,3-pyridinedicarboxylic acid ligand. The structures of the compounds were characterized by elemental analysis (E.A), infrared spectroscopy (FT-IR), nuclear magnetic resonance (NMR), ultraviolet visible spectroscopy (UV-vis) and single crystal X-ray diffraction analysis. Crystal structure reveal that the coordination number of 2,3-pyridinedicarboxylic acid ligand was 6, and each ligand is connected to a metal ion. The metal is connected to the ligands by n-metal and o-metal bonds, and the structural diversity is controlled by the core metal ions. The IC50 test showed that complex Ⅰ-Ⅵ had low to medium activity on human tumor cell line SMMC-7721, and complex Ⅵ had the best activity, with IC50 value of 21.80.
Keywords: 2,3-Pyridinedicarboxylic acid; Metal complex; Crystal structure; Antitumor activity
Introduction
Due to their distinctive structures and excellent properties, metal complexes have been widely used in catalysis [1], biosensing [2], gas storage [3] and capture [4] and molecular recognition and separation [5], and they exhibit antifungal [6], antituberculous [7], analgesic [8] and anticancer [9] activities. Organic carboxylic acids are widely used as ligands in the design of metal-organic complexes because they can bridge multiple metal ions using various coordination modes, such as monodentate coordination and bidentate chelation [10]. In 1995, Yaghi's [11] research group first used an organic carboxylic acid as a ligand, reacted with Co metal to produce metal coordination compounds and proposed the concept of metal-organic framework materials. Since then, carboxylate molecules have been studied intensively because they offer different design capabilities based on coordination with metal ions [10]. During the synthesis of metal complexes, there are many factors affecting the crystal growth, including temperature, solvent and PH value of the system, among which the properties of metal ions and organic ligands themselves are the key factors in the crystal synthesis process [12-14]. It is found that appropriate metal ions and organic ligands not only directly affect the formation of complexes, but also play a key role in the spatial structure of the products, so the selection of appropriate ligands is the primary task [15]. Carboxylate oxygen and N atoms in N-heterocyclic aromatic carboxylate ligands can be metal-coordinated and can also act as multiple proton donors and acceptors, thus forming an interesting network structure [16]. As a bridging ligand, 2,3-pyridine dicarboxylic acid can react with transition metals. Due to the asymmetric coordination environment of 2-site and 3-site carboxylic groups in the pyridine ring, it is a good carboxylate ligand with different coordination capabilities and flexible and diverse coordination modes [17-18].
The type of the central metal of the metal complex can affect the inhibition effect of the compound as an anticancer drug, and the organic ligand coordinated with metal ions will also have an important impact on its biological activity [19]. The adverse effects of metals may be reduced by coordination with organic ligands, and in this regard, 2, 3-pyridinic acid complexes may be a favorable choice [20-21]. Therefore, our research team successfully synthesized and characterized a series of copper (Ⅱ), zinc (Ⅱ), cobalt (Ⅱ) and nickel (Ⅱ) metal complexes by one-pot synthesis using 2, 3-pyridine dicarboxylic acid as ligands. In addition, compared with other synthesis methods, our synthesis method has the advantages of high yield, low toxicity, no smoke, environmental friendliness and high cost performance.
In this paper, we report six metal complexes produced with the one-pot synthesis shown in (Scheme 1), [CuC20H20N6O10] (Ⅰ), [ZnC7H9NO7] (Ⅱ), [CoC21H18N3O15] (Ⅲ), [CoC14H10N2O10] (Ⅳ), [NiC14H10N2O10] (Ⅴ) and [ZnC14H10N2O10] (Ⅵ), which were characterized by single crystal X-ray diffraction, UV‒vis, FT-IR and E.A. In 2013, Shankar's group successfully synthesized the complex (Ⅰ)-[CuC20H20N6O10] for the first time. In the reaction process, Shankar's group first reacted 2, 3-pyridinic acid with metal salts, and finally added imidazole molecule. Our group fully reacted carboxylic acid molecule with imidazole before adding metal salts for the reaction, and tested the anticancer activity of the resultant complex [22]. In 2018, the Terenti group successfully synthesized the complex (Ⅲ)-[CoC21H18N3O15] for the first time, during the reaction process, Terenti’s group added (NH4)2S2O8 (ammonium persulfate), which is highly oxidizing and corrosive, and is harmful to the environment and the human body. Our group directly synthesized the complex (Ⅲ) using one-pot method, which has the advantages of high yield, low toxicity, and cost-effective, and we used nontoxic and non-hazardous solvents and reagents in all of the reaction process, and synthesized a high yield of the complex (Ⅲ) [23]. In 2007, Baruah's team synthesized the complex [Co(LH)2(H2O)2 (LH2=2,3-pyridindicarboxylic acid) for the first time. Compared with the complex (Ⅳ)-[CoC14H10N2O10] synthesized by our team, the 3-carboxyl group of the complex synthesized by Baruah's team did not deprotonate. The complex (Ⅳ) synthesized by our team was completely deprotonated, and we tested its anticancer activity [24]. In 2011, the Baruah group again synthesized the complex [Co(LH)2(H2O)2, which is very similar to our complex (Ⅴ)-[NiC14H10N2O10], but with different central metal ions [25]. In 2020, Soudani's team synthesized the monodeprotonated complex [Zn(2,3-pdcH)2(H2O)2] (2,3-pdcH =2,3-pyridinic acid) using the reaction of 2, 3-pyridinic anhydride and zinc dichloride, using the same reaction method as our team. However, our team has tested the anti-cancer activity of complex (Ⅵ)-[ZnC14H10N2O10], and the anti-cancer activity is good [26]. Complex (Ⅱ)-[ZnC7H9NO7] is a novel metal complex synthesized by our team and has good anticancer activity.
Cancer is the most serious disease nowadays, and scholars are actively searching for drugs that can treat cancer, while our synthetic complexes have good cytotoxicity to lung cancer cells, but negligible toxicity to normal cells.
Experimental
General Experimental Details
2,3-Pyridinedicarboxylic acid, Cu(OAC)2•H2O, Zn(OAC)2•2H2O, Co(NO3)2•6H2O, CoCl2•6H2O, NiCl2•6H2O and ZnCl2 were purchased from Acros. The reagents were obtained from commercial sources. All reactions were performed in flame-dried glassware under normal atmospheric pressure. Nuclear magnetic resonance (NMR) spectra were acquired on a 500 MHz Bruker Advance III spectrometer. Infrared spectra were recorded on a Mattson Galaxy Series FTIR 3000 spectrometer; peaks are reported in cm–1. Elemental analysis was performed on a VARIO ELIII elemental analyser. The crystal structures were determined by using a Gemini S Ultra diffractometer. The chemical shifts of 1H and 13C NMR
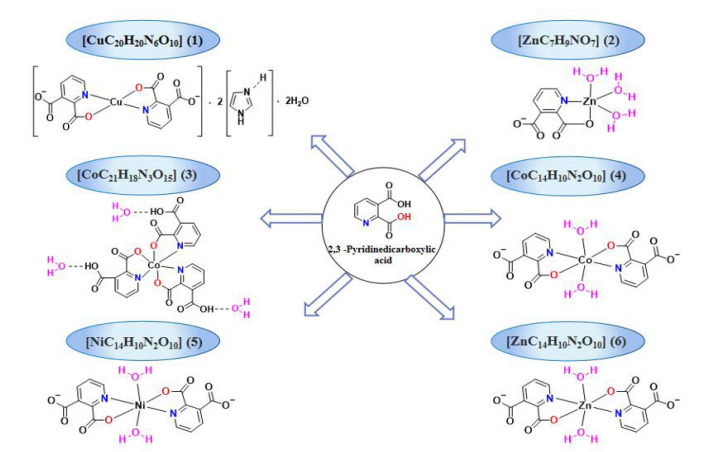
Figure 1: Synthetic routes to complexes (I)-(Ⅵ).
reported in ppm, with referenced to 7.26 ppm of CDCl3; the concentration of DMSO-d6 is 2.50 ppm. Use the following abbreviations: s = singlet, d = doublet, t = triplet, q = quartet, m = multistate. The melting points were determined by the Yanaco miniature melting point device MP-J3 and the SANSYO Melting point instrument (SMP-500).
Cytotoxicity assay
The human hepatocellular carcinoma cell line SMMC-7721 was used as the research object to carry out in vitro killing experiment. These cell lines were obtained from ATCC (Manassas, VA, USA). The cells were cultured in RMPI-1640 or DMEM (Biological Industries, Kibbutz Beit Haemek, and Israel) supplemented with 10% foetal bovine serum (Biological Industries) at 37 °C in a humidified atmosphere with 5% CO2. Cytotoxicity was tested by MTS method (Promega, Madison, WI, USA). In this project, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) was used as the research object to evaluate its inhibitory effect on tumor cells by testing its killing effect on tumor cells. In short, 96 holes in the cells culture dish, each hole is planted with a batch of cells. Culture at 37 ℃ for 12 hours and add 100 μM of the substance to be measured. After incubation for 48 h, the growth of the cells was detected by MTS method. Compounds with 50% growth inhibition rate were further evaluated using cisplatin and paclitaxel (Sigma, St. Louis, MO, USA) as positive controls at concentrations of 0.064, 0.32, 1.6, 8, 40 and 100 μM.
General Procedure for the Syntheses of Complexes (I)-(Ⅵ)
The ligand and metal salts (molar ratio of 1:1) were heated and refluxed for 48 h, then filtration was conducted immediately after the reaction, and the filtrate was retained for slow volatilization. The metal-ligand complexes were successfully synthesized by reacting the 2,3-pyridinedicarboxylic acid ligand with Cu(OAC)2•H2O, Zn(OAC)2•2H2O, Co(NO3)2•6H2O, CoCl2•6H2O, NiCl2•6H2O and ZnCl2, and the resulting crystals were analysed and characterized by X-ray diffraction, FT-IR, 1H NMR, 13C NMR, UV‒vis and E.A. The first key was to find the right ligands, and then the ligands and the corresponding metal salts were reacted together. At the end of the reaction, filtration was carried out, and a suitable solvent for crystal precipitation was found. The selection among available solvents such as anhydrous methanol, ethanol, chloroform, etc., was the most critical step. The crystals were frozen in a refrigerator if these substances did not precipitate at room temperature.
Synthesis of the 2,3-pyridine dicarboxylate copper complex (Ⅰ)
2,3-Pyridine dicarboxylic acid (0.167 g, 1 mmol) and imidazole (0.068 g, 1 mmol) were weighed into a 100 ml round-bottomed flask and stirred with 20 ml of deionized water to dissolve them; Cu(OAC)2•H2O (0.200 g, 1 mmol) was added to the above solution and heated at 90 °C for 48 h. After the reaction was complete, the solution was filtered into a 50 ml beaker while it was hot. The beaker was covered with a layer of plastic wrap that was pierced with a fine needle to allow the solvent to evaporate naturally at room temperature. After 3 days, blue crystals had separated out, which were suitable for X-ray single-crystal analysis. The yield was 70%, m. p. 260-265 °C. IR (KBr, υ, cm−1): 3430 (-O-H), 3140 (-N-H), 3070 (-C-H), 2970, 2540, 1950, 1780, 1650 (-C=O), 1580 (-C=C), 1430 (-C=N), 1350 (-C-C), 1270, 1230, 1175, 1160, 1120 (-C-O), 1070, 910, 867, 836, 731, 696, 634 (-Cu-O), 557 (-Cu-N). For [CuC20H20N6O10] anal. calcd., %: N, 14.797; C, 42.29; H, 3.537. Found, %: N, 15.29; C, 41.87; H, 3.911.
Synthesis of the 2,3-pyridine dicarboxylate zinc complex (Ⅱ)
Complex (II) was synthesized according to the general procedure using 2,3-pyridine dicarboxylic acid (0.167 g, 1 mmol) and Zn(OAC)2•H2O (0.220 g, 1 mmol). After 3 days of natural evaporation, white crystals appeared at the walls of the vessel, which were suitable for X-ray single-crystal analysis. The yield was 70%, m. p. 320-325 °C. IR (KBr, υ, cm−1): 3380 (-O-H), 3280 (-C-H), 2530, 2250, 2000, 1960, 1910, 1630 (-C=O), 1590, 1560 (-C=C), 1460 (-C=N), 1410, 1380 (-C-C), 1280, 1240, 1160, 1110 (-C-O), 1060, 885, 831, 713, 676, 619 (-Zn-O), 549 (-Zn-N). For [ZnC7H9NO7] anal. calcd., %: N, 4.63; C, 27.79; H, 3.665. Found, %: N, 4.98; C, 28.07%; H, 3.465%.
Synthesis of the 2,3-pyridine dicarboxylate cobalt complex (Ⅲ)
Complex (Ⅲ) was synthesized according to the general procedure using 2,3-pyridine dicarboxylic acid (0.167 g, 1 mmol) and Co(NO3)2•6H2O (0.247 g, 1 mmol). After 2 days of natural evaporation, amaranth crystals appeared at the walls of the vessel, which were suitable for X-ray single-crystal analysis. The yield was 90%, m. p. 240-245 °C. IR (KBr, υ, cm−1): 3500 (υO-H), 3440 (-O-H), 3100 (-C-H), 2930, 2630, 2520, 1940, 1760, 1720, 1680 (-C=O), 1600 (-C=C), 1460 (-C=N), 1340 (-C-C), 1280, 1230, 1110 (-C-O), 1010, 839, 696, 620 (-Co-O), 567 (-Co-N). 1H NMR (600 MHz, DMSO) δ 8.56-8.57 (d, 1H), 8.52-8.43 (d, 1H), 8.40-8.45 (t, 3H), 8.04-8.09 (m, 2H), 7.86-7.88 (t, 1H), 7.65-7.66 (d, 1H); 13C NMR (151 MHz, DMSO) δ 170.82, 170.75, 169.99, 166.69, 166.59, 166.41, 151.16, 150.76, 150.01, 148.45, 147.43, 147.13, 140.79, 140.11, 139.90, 136.11, 135.68, 135.54, 131.5, 130.61, 130.52. For [CoC21H18N3O15] anal. calcd., %: N, 6.87; C, 41.26; H, 2.968. Found, %: N, 7.22; C, 41.47; H, 3.390.
Synthesis of the 2,3-pyridine dicarboxylate cobalt complex (Ⅳ)
Complex (Ⅳ) was synthesized according to the general procedure using 2,3-pyridine dicarboxylic acid (0.167 g, 1 mmol) and Co(Cl)2•6H2O (0.238 g, 1 mmol). After 2 days of natural evaporation, orange red crystals appeared at the walls of the vessel, which were suitable for X-ray single-crystal analysis. The yield was 85%, m. p. 210-215 °C. IR (KBr, υ, cm−1): 3410 (-O-H), 3090 (-C-H), 1680 (-C=O), 1560 (-C=C), 1440 (-C=N), 1380 (-C-C), 1270, 1170, 1100 (-C-O), 878, 831, 758, 687, 656 (-Co-O), 592 (-Co-N). For [CoC14H10N2O10] anal. calcd., %: N, 6.12, C, 39.37, H, 3.06%. Found, %: N, 6.56, C, 39.33, H, 3.198.
Synthesis of the 2,3-pyridine dicarboxylate nickel complex (Ⅴ)
Complex (Ⅴ) was synthesized according to the general procedure using 2,3-pyridine dicarboxylic acid (0.167 g, 1 mmol) and Ni(Cl)2•6H2O (0.238 g, 1 mmol). After 3 days of natural evaporation, baby blue crystals appeared on the walls of the vessel, which were suitable for X-ray single-crystal analysis. The yield was 78%, m. p. 230-235 °C. IR (KBr, υ, cm−1): 3420 (-O-H), 3100 (-C-H), 1680 (-C=O), 1560 (-C=C), 1440 (-C=N), 1370 (-C-C), 1270, 1180, 1110 (-C-O), 875, 821, 759, 690, 658 (-Ni-O), 598 (-Ni-N). For [NiC14H10N2O10] anal. calcd., %: N, 6.32; C, 37.96; H, 2.730. Found, %: N, 6.65; C, 38.37; H, 3.062.
Synthesis of the 2,3-pyridine dicarboxylate zinc complex (Ⅵ)
Complex (Ⅵ) was synthesized according to the general procedure, with 2,3-pyridine dicarboxylic acid (0.167 g, 1 mmol) and Ni(Cl)2•6H2O (0.136 g, 1 mmol) dissolved by stirring in 10 ml of deionized water and 10 ml of anhydrous ethanol. After 3 days of natural evaporation, white crystals appeared on the walls of the vessel, which were suitable for X-ray single-crystal analysis. The yield was 83%, m. p. 240-245 °C. IR (KBr, υ, cm−1): 3440 (-O-H), 3100 (-C-H), 1690 (-C=O), 1560 (-C=C), 1440 (-C=N), 1390 (-C-C), 1270, 1180, 1110 (-C-O), 872, 823, 757, 691, 660 (-Zn-O), 590 (-Zn-N). 1H NMR (600 MHz, DMSO) δ 8.49-8.59 (t, 4H), 7.82–7.84 (m, 2H); 13C NMR (151 MHz, DMSO) δ 166.75, 166.73, 166.35, 166.34, 148.92, 148.90, 148.87, 146.88, 142.20, 142.14, 132.35, 127.96, 127.95, 88.01. For [ZnC14H10N2O10] anal. calcd., %: N, 6.96, C, 37.80, H, 3.506. Found, %: N, 6.52, C, 38.17, H, 3.222.
X-ray structure
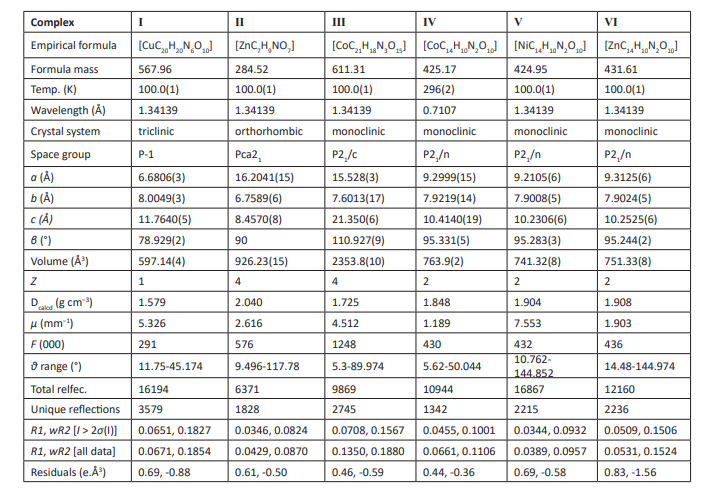
X-ray diffraction data for complexes (I)-(Ⅵ) were collected at room temperature using graphite-monochromatic Mo Kα radiation (λ= 0.71073 Å) on an Oxford Diffraction Geminis diffractometer. Structure solutions and refinements for complexes Ⅰ-Ⅵ were carried out with the programs SHELXT [27] and SHELXL-2018/3 [28], respectively. MERCURY [29] was employed for molecular graphics and OLEX2 [30]. In (I)-(Ⅵ), anisotropic purification of non-hydrogen atoms and isotropic restriction purification of hydrogen atoms are performed. Table 1 summarizes the crystallography and refining parameters of the (I)-(VI) complexes. The selected bond lengths and bond angles are listed in Table 1 respectively, while the hydrogen bonds of complex I-IV are listed in Table 2 respectively.
Table 1: Cell parameters and crystallographic data for complexes (I)–(Ⅵ).
Results and discussion Materials and methods
Complexes (I)-(IV) were synthesized by a one-pot method, and the synthetic routes are shown in (Figure 2). Complexes (I)-(IV) were synthesised in deionized water or anhydrous ethanol from the 2,3-pyridine dicarboxylic acid ligand and different metal salts, such as Cu(OAC)2•H2O, Zn(OAC)2•2H2O, Co(NO3)2•6H2O, CoCl2•6H2O, NiCl2•6H2O, ZnCl2 (1:1 eq.) while heated at reflux 48 h. The solutions were filtered while hot so the solvents evaporated naturally at room temperature to precipitate crystals. The products were characterized by XRD, FT-IR, 1H NMR, 13C NMR, UV‒vis and E.A.
Crystal structure analyses of complexes (I)-(IV)
The crystal structures and stereograms of complexes (I)-(IV) are shown in (Figure 1a),
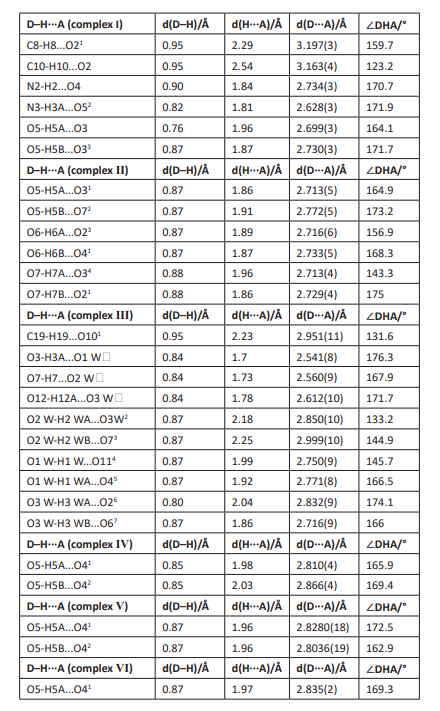
Table 2: Hydrogen bond lengths (Å) and bond angles (°) for complexes (I)-(Ⅵ).
coordination environment is shown for each metal atom in complexes (I)-(IV) in (Figure 1b). Under these experimental conditions, the space groups of complexes (I)/(Ⅱ)/(Ⅲ) were P-1/Pca21/P21/c, respectively, and those of complexes (Ⅳ)/(Ⅴ)/(Ⅵ) were P21/n (Table 1). For the mononuclear metal complexes (I)-(IV), there was only one metal ion. For complex (I), the symmetric unit contained a copper ion, two 2,3-pyridine dicarboxylic acid molecules, two imidazole molecules and two water molecules. For complex (Ⅱ), the asymmetric unit comprised one zinc ion, one 2,3-pyridine dicarboxylic acid molecule and three water molecules. For complex (Ⅲ), the asymmetric unit contained one cobalt ion, three 2,3-pyridine dicarboxylic acid molecules and three water molecules. Complexes (Ⅳ)- (Ⅵ) were all consistent except for the metal ions, and they contained two 2,3-pyridine dicarboxylic acid molecules and two water molecules.
In the crystal structure of complex (I), the coordination environment of the central metal Cu1 exhibited a hexagonal regular octahedron (Fig. 2b). The copper atom was bonded to the two nitrogen atoms (N1, N11) in the two different 2,3-pyridine dicarboxylic acid ligands and two oxygen atoms (O1, O11) and the other two oxygen atoms (O42, O43) of the two pyridine dicarboxylate ligands extended into space. The lengths of the six bonds were dCu1-N1 / N11= 1.972(2) Å, dCu1-O1 / O11 = 1.9658(18) Å, dCu1-O42 / O43 = 2.4351(18) Å. With the copper atoms defined as the centre, the cis angles were 91.69(8)°, 88.31(8)°, 96.93(8)°, 83.07(8)°, 95.96(8)° and 84.04(7)°; the trans angles were all 180°, which was similar to other analogues [31, 32]. In the crystals, the pyridine rings belonging to adjacent complexes were stacked in a parallel displacement pattern, and there was a three-dimensional hydrogen bond network formed by waters and the uncoordinated imidazole molecules (Table S1 and Fig. S1).
In the crystals of complex (Ⅱ), the coordination environment of the Zn1 central metal showed an irregular octahedral geometry with hexagonal coordination (Fig. 2b). The zinc atom was bonded with a nitrogen atom (N1) from the 2,3-pyridine dicarboxylic acid ligand, an oxygen atom from a deprotonated 2-carboxylic acid group (O1), three oxygen atoms from the three coordinated water molecules (O5, O6, O7) and the oxygen atom (O41) from another 2,3-pyridine dicarboxylate ligand. The six key lengths were dZn1-N1 = 2.139(4) Å, dZn1-O1 = 2.063(3) Å, dZn1-O41 = 2.102(4) Å, dZn1-O5 = 2.072(3) Å, dZn1-O6 = 2.082(4) Å, and dZn1-O7 = 2.230(3) Å, which confirmed the formation of irregular octahedra. During crystallization, hydrogen bonds were formed between the hydrogen atoms of the water molecules and the oxygen atoms of the 2,3-pyridine dicarboxylate ligands [33]. The hydrogen bonding interactions between the 2,3-pyridine dicarboxylic acid ligands and water molecules and the p-p stacking interactions between the water molecules may have led to the stacking arrangement of the molecules in the crystals [34] (Table S1 and Fig. S1).
In complex (Ⅲ), the coordination environment of the Co1 central metal exhibited a hexagonal twisted octahedral geometry (Fig. 2b). The cobalt atom was coordinated by three nitrogen atoms (N1, N2, N3) and three oxygen atoms (O1, O5, O9) from three different 2,3-pyridine dicarboxylic acid ligands. The longest axis comprised the Co1-N1 and Co1-N2 bonds, and their lengths were 1.939(7) Å and 1.911(7) Å, respectively. The shortest axis comprised the Co1-O1 and Co1-O9 bonds, with lengths of 1.876(6) Å and 1.877(6) Å, respectively. The last axis comprised the Co1-O5 and Co1-N3 bonds, with lengths of 1.901(6) Å and 1.899(7) Å, respectively; these were comparable to those of reported analogues [35, 36]. In the crystal, each 2,3-pyridine dicarboxylic acid ligand was coordinated with Co1 via bidentate chelation involving the pyridine nitrogen atom and the oxygen atom to form a five-membered chelate ring. The remaining protonated carboxyl groups did not undergo coordination but formed intramolecular hydrogen bonds with the three water molecules, resulting in a three-dimensional hydrogen bond network [37] (Table S1 and Fig. S1).
In complex (Ⅳ), the Co1 central metal was coordinated with two nitrogen atoms (N1, N11) from two different 2,3-pyridine dicarboxylic acid ligands, two oxygen atoms from the deprotonated 2-carboxylic acid groups (O1, O11), and two oxygen atoms (O5, O51) from two coordinated water molecules (Fig. 2b). Therefore, an octahedral geometry was adopted. The bond angles O1-Co1-O11, O51-Co1-O5, and N11-Co1-N1 were all 180°, which confirmed that all atoms were arranged in the same planes relative to the metal centre. The longest axis comprised the Co1-O5 and Co1-O51 bonds, both of which had lengths of 2.154(3) Å. The shortest axis comprised the Co1-O1 and Co1-O11 bonds, both of which had lengths of 2.043(2) Å. The last axis comprised Co1-N1 and Co1-N11, both of had lengths of 2.073(3) Å, which were comparable to those of analogues [38]. In the crystal packing structure, the hydrogen bonds between the ligands and the water molecules showed bond spacings of 2.810(4) Å and 2.866(4) Å (O5-H5A-O41 and O5-H5B-O42, respectively), and adjacent molecules were connected to form a three-dimensional supramolecular structure [17] (Table S1 and Fig. S1).
In complex Ⅴ, the coordination pattern and ligands were similar to those of complex Ⅳ, except that the central metal was different from that of complex Ⅳ (Fig. 2b). The six bond lengths were dNi1-N1/N11= 2.0277(14) Å, dNi1-O1/O11 = 2.0312(12) Å, and dNi1-O5/O51 = 2.1120(12) Å, which were similar to those of some analogues. Because the crystal structure was similar to that of complex (Ⅳ), no further discussion is provided. The hydrogen-bonded ligands and water molecules interacted to link the adjacent molecules and form a three-dimensional supramolecular structure (Table S1 and Fig. S1).
In complex (Ⅵ), the coordination mode and ligands were similar to those of complexes (Ⅳ) and (Ⅴ), except that the central metal was different from those of complexes (Ⅳ) and (Ⅴ) (Fig. 2b). The six bond lengths were dZn1-N1/N11 = 2.0679(16) Å, dZn1-O1/O11 = 2.0598(14) Å, and dZn1-O5/O51 = 1.833(14) Å, which were similar to those of analogues. Because the crystal structure was similar to those of complexes (Ⅳ) and (Ⅴ), no further discussion is provided. Hydrogen bonding between the ligands and water molecules linked the adjacent molecules and formed a three-dimensional supramolecular structure (Table S1 and Fig. S1).
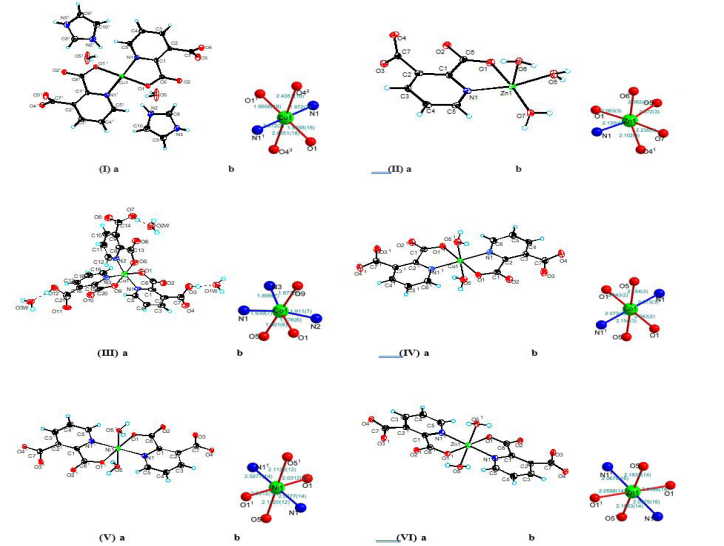
Figure 2a: Complexes (I)-(Ⅵ) shown as ORTEP molecular structures with 30% probabilities for the thermal ellipsoids.
2b: Coordination environments of the metal atoms in complexes (I)-(Ⅵ).
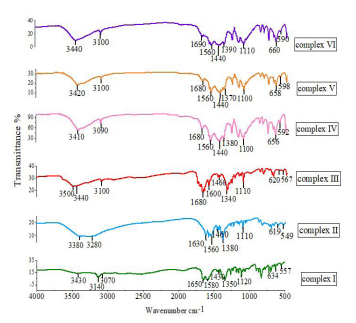
Figure 3: IR spectra of complexes (I)-(Ⅵ) in the 4000-500 cm-1 region.
Table 3: Cytotoxicities of complexes (I)-(Ⅵ) to the human tumour cell line SMMC-7721.
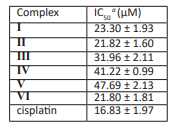
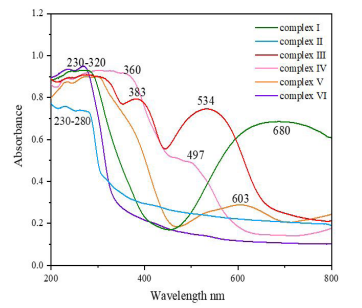
Figure 4: UV‒vis spectra of complexes (I)-(Ⅵ) in the range 200-800 nm.
IR analyses of complexes I-VI
IR analyses of complexes I-VI
The IR spectra showed many peaks for all of the complexes (Fig. 3). In infrared spectroscopy, it is usually found that the vibrations of O-H groups are most affected by the environment. Therefore, the O-H group shows a distinct absorption peak in the infrared spectrum. Generally, the O-H stretching vibrations give peaks within the range 3750–3000 cm-1, and when there is no hydrogen bonding of the free O-H groups, they exhibit sharp absorption peaks. When the O-H group has a hydrogen bonding O--H/O interaction, a broad absorption peak is formed, such as with H2O [39]. In the complexes (I)–(Ⅵ) studied here, obvious absorption peaks were observed in the infrared spectra at 3440-3380 cm-1, and all of them were broad peaks; this indicated that there were hydrogen bond O--H/O interactions in the complexes, that is, the complexes (I)–(Ⅵ) contained waters of crystallization. In complex (Ⅲ), a broad absorption peak also appeared at 3500 cm-1, which was the infrared absorption peak of a carboxyl group O-H. The stretching vibrations of N-H groups generate peaks in the range 3550–3250 cm-1. When an associated N-H group is present, its stretching vibrational peak moves to lower energy, and the displacement is generally approximately 100 cm-1 [40]. In complex (I), the absorption peak at 3140 cm-1 was obvious, and it indicated the presence of associated N-H groups. In the (Ⅰ)-(Ⅵ) complex, the C-H expansion vibration of unsaturated carbon chains in the aromatic ring has different absorption peaks between 3300-3000 cm-1, while the absorption peak between 1600-1560 cm-1 is the expansion vibration of C= C in the aromatic ring, which proves that it has an aromatic structure [41]. In complexes (I)–(Ⅵ), the C=O stretching vibrations generated peaks within the range 1690–1630 cm-1, and the absorption peaks for C=N, C-O and C-C vibrations were found at 1460–1000 cm-1. Due to the interactions between the metals and ligands, peaks for the metal-nitrogen tensile vibrations were observed within the range 660–600 cm-1 and those for the metal-oxygen vibrations were in the range 600–500 cm-1 [42, 43] (Fig. S2).
UV‒vis spectral analyses of complexes (I)-(Ⅵ)
The solid-state UV-vis. In order to determine their light absorption properties, the spectra of complex Ⅰ-VI in the band of 200-800 nm were recorded (Fig. 4). Complexes Ⅰ/Ⅱ/Ⅵ at 230-280 nm, complexes Ⅲ/Ⅴ at 230-320nm and complex Ⅳ at 230-360nm all have a wide absorption peak. This may be caused by the n→σ* transition of the C-O/C-N groups in the complex or by the n→π* transition of the C=O group. It is also possible that the intrinsic absorption peaks of the aromatic molecules in the complexes overlapped with the π→π* transitions. The absorption peak seen at 383 nm for complex (Ⅲ) was produced by an n→π* transition of the COOH group [44, 45]. Transition metal ions have degenerate d orbitals, and when dipolar molecules such as H2O or anions such as COO- and CN- are coordinated to the transition metal ions and form certain geometric shapes, these degenerate d orbitals are split into energy levels with different energies, and most of the gaps correspond to the visible region of the spectrum [46]. The central metal of complex (I) is Cu2+, which has a deep blue color and has the strongest absorbance, so it has the broadest absorption peak at 680 nm, which is caused by the d→d* transitions of Cu2+. The central metals of complexes Ⅲ/Ⅳ are both Co2+, but the color of complex Ⅲ is orange-red, while the color of complex Ⅳ is light pink, so the absorption peak of complex Ⅲ at 534 nm is stronger than that of complex Ⅳ at 497 nm, which is caused by the d→d* transitions of Co2+. The central metal of complex Ⅴ is Ni2+, and its complex color is light blue with weak absorbance, so there is a weak absorption peak at 603 nm, which is caused by the d→d* transitions of Ni2+. For complexes (II)/(VI), the central metal is Zn2+, which formed colourless complexes that did not absorb visible light, so no absorption peaks were found [47].
Cytotoxicity assays of complexes (I)-(Ⅵ)
Table 3 shows the IC50 values of the six compounds and cisplatin, which were tested and calculated by the Reed and Muench methods [48]. The carboxylic acid metal complexes (I)-(Ⅵ) showed low to moderate activities against the human tumour cell line SMMC-7721. Complex (Ⅵ) showed the best activity, and the IC50 value was 21.80.
During the study, the physicochemical characteristics of the ligands and metal ions were considered to understand the structure-activity relationships of the tested metal complexes. Structurally, we believe that the central ion of the metal coordination sphere, the properties and mode of coordination of the ligand, and the properties of the auxiliary ligand are related to the observed activity [49]. For example, the activities of the zinc complexes (Ⅱ) and (Ⅵ) were generally higher than that of the Cu complex (Ⅰ), the activity of Cu complex (Ⅰ) was significantly greater than those of the Co complexes (Ⅲ) and (Ⅳ) and the Ni complex (Ⅴ). The anticancer activity of the Zn complex (Ⅵ) was better than that of complex (Ⅱ) because there were N and O atoms coordinated from the two pyridine carboxylic acid ligands in complex (Ⅵ), which was the most important factor for the pyridine carboxylic acid metal complexes [50]. A further study of the coordination environments of metal atoms and their activities is under way in our laboratory.
ɑCytotoxicities were expressed as IC50 values for each cell line, i.e., the complex concentration that caused a 50% reduction relative to the untreated SMMC-7721 cells as determined with the SRB assay. Cisplatin was used as the experimental control.
Conclusions
The complexes [CuC20H20N6O10] (Ⅰ), [ZnC7H9NO7] (Ⅱ), [CoC21H18N3O15] (Ⅲ), [CoC14H10N2O10] (Ⅳ), [NiC14H10N2O10] (Ⅴ) and [ZnC14H10N2O10] (Ⅵ) were synthesized by a one-pot method. The crystal structures of these complexes were determined by X-ray diffraction and characterized by E. A, FT-IR, NMR and UV‒Vis. The single-crystal X-ray data showed that the complexes 1-6 are all mononuclear metal complexes with only one central metal ion and all have coordination number 6, forming an octahedral configuration. Complexes (I)–(Ⅵ) showed low to moderate activities against the human tumour cell line SMMC-7721. Complex (Ⅵ) showed the best activity, and the IC50 was 21.80 μM. It was observed that the anticancer activities of these complexes depended on the type of metal ion, cell line and geometry of the corresponding molecule. These useful results provide the impetus for design and development of novel therapeutic drug-like molecules. The synthesized complexes have been tested in catalytic reactions and provided excellent results. Studies of the catalytic activities of complexes (I)-(Ⅵ) in different organic reactions are currently underway.
CCDC-2218325 (Ⅰ),-2218327 (Ⅱ),-2218326 (Ⅲ),-2218396 (Ⅳ),-2218323 (Ⅴ),-2218324 (Ⅵ) are supplementary crystallographic data for this paper. This material is freely available from Cambridge's Cambridge database at: www.ccdc.cam.ac.uk/data_request/cif.
Supporting Information: Bond lengths and angles, crystal stacking, infrared data.
Acknowledgements: The project was funded by Hefei University of Science and Technology and the Key Laboratory of Photochemistry and Plant Resources in Western China.
Declarations
Conflict of interest: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- V.Jornet-Molla, C. Dreessen, F.M. Romero, Inorg. Chem. 105 2021; 60: 72-10584.
- I.Majumder, P. Chakraborty, R. Alvarez, M. Gonzalez-Diaz, R. Pelaez, Y. Ellahioui, et al. Omega. 2018; 3: 13343-13353.
- Z.N. Mahmood, M. Alias, G. A.-R. El-Hiti, D.S. Ahmed, E. Yousif, Korean J. Chem. Eng. 2021; 38: 179-186.
- Y.C. Lin, C.L. Kong, Q.J. Zhang, L. Chen, Adv. Energy. Mater. 2016; 7: 1601296-1601325.
- M.Yan, F. Jiang, H.T. Fei, F.G. Ma, J.Y. Yan, Y.L. Wu, J. Colloid. Inter. Sci. 2022; 606: 696-708.
- W.X. Liu, Y.K. Qin, S. Liu, R.G. Xing, H. Yu, X.L. Chen, K. Li, P. Li, Sci. Rep. 2018; 8: 4845-4855.
- G.J. Kharadi, J.R. Patel, B.Z. Dholakiya, Appl. Organomet. Chem. 2010; 24: 821-827.
- M.M.U. Mazumder, A. Sukul, S.K. Saha, Dhaka. Univ. J. Pharm. Sci. 2016; 15: 89-96.
- Q.Umar, Y. Huang, A. Nazeer, H. Yin, J.C. Zhang, M. Luo, X.G. Meng, RSC. Adv. 2022; 12: 32119-32128.
- S.W. Zhang, W. Shi, P. Cheng, Coordin. Chem. Rev. 2017; 352: 108-150.
- O.M. Yaghi, G.G. Li, H.L. Li, Nature. 1995; 378: 704-706.
- P.Hayati, S.S. Garcra, A. Gutierrez, DR Molina, A Morsali, A.R. Reazvani, Ultrason. Sonochem. 2018; 42: 320-326.
- Causero, G. Ballman, J. Pahl, C. Farber, J. Inteman, S. Harder, Dalon. Trans. 2017; 46: 1822-1831.
- Y. Yao, Q.B. chen, Y.J. Sheng, A.T. Kai, HL. Liu, CrystEngComm. 2017; 19: 5835-5843.
- A.Hoque, J. Benet-Buchholz, A. Llobet, C. Gimbert-Surinach, ChemSusChem. 2019; 12: 1949-1959.
- H. Wang, Z.G. Li, H.Q. Jia, N.H. Hu, J.W. Xu, CrystEngComm. 2009; 11: 292-297.
- K. Gangu, S. Maddila, S.B. Mukkamala, S.B. Jonnalagadda, J. Mol. Struct. 2017; 1143: 153-184.
- Semerci, O.Z. Yesilel, S. Keskin, C. Darcan, M. Tas, H. Dal, CrystEngComm. 2013; 15: 1244-1256.
- Abdolmaleki, A.Aliabadi, M. Ghadermazi, Inorg. Chem. Acta. 2022; 542: 121152-121162.
- Aliabadi, M. Hakimi, F. Hosseinabadi, E. Motieiyan, V.H.N. Rodrigues, M. Ghadermazi, D. et al. J. Mol. Struct. 2021; 1223: 129005-129016.
- Aliabadi, M. Zangeneh, A. Izadi, M. Badzohre, M. Ghadermazi, D. Marabello, et al. J. Mol. Struct. 2022; 1247: 131327-131336.
- Shankar, B. Das, J.B. Baruah, ChemPubSoc. 2013; 36: 6147-6155.
- Terenti, M. Ferbinteanu, A. Lazarescu, Chem. J. Mod. 2018; 13: 24-29.
- M. Baruah, A. Karmakar, J.B. Baruah, Polyhedron. 2007; 26: 4518-4524.
- B. Baruah, J.Chem. Sci. 2011; 123: 123-129.
- Soudani, M. Hajji, J.X. Mi C. Jelsch, F. Lefebvre, T. Guefel, C.B. Nasr, J. Mol. Struct. 2020; 1199: 127015-127025.
- A. Amer, J. Heterocyclic Chem. 2008; 55: 297-302.
- V. Dyachenk, V.D. Dyachenko, P.V. Dorovatovsky, V.N. Khrustalev, V.G. Nenajdenko, Russ. J. Org. Chem. 2020; 56: 974-982.
- Purushothaman, K. Thanigaimani, S. Arshad, S. Silambarasan, I.A. Razak, K.M.S. Ali, Acta. Cryst. 2014; 70: 0812-0813.
- L. Luttle, W.J. Middleton, D.D. Coffman, V.A . Engelhardt, G.N. Sausen, J. Am. Chem. Soc. 1958; 80: 2832-2839.
- Morikawa, T. Yamada, H. Kitagawa, Chem. Lett. 2009; 38: 654-656.
- Yamada, S. Morikawa, H. Kitagawa, Bull. Chem. Soc. Jpn. 2010; 83, 42-48.
- A. Saghatforoush, L. Valencia, F. Chalabian, S. Ghammamy, L.Z. Khaledi, J. Coord. Chem. 2011; 64: 3311-3322.
- Mandado, M.N.D.S. Cordeiro, J. Comput. Chem. 2010; 31: 2735-2745.
- A.S. Goher, A.A. Youssef, F.A. Mautner, Polyhedron. 2006; 25: 1531-1536.
- T. Colak, G. Pamuk, O.Z. Yesilel, F. Yilmaz, O. Buyukgungor, J. Chem. Crystallogr. 2012; 42: 76-82.
- Q. Chai, L. Zhou, H.B. Zhang, K.H. Mao, H.S. Zhang, New. J. Chem. 2019; 43: 12417-12447.
- M. Baruah, A. Karmakar, J.B. Baruah, Polyhedron. 2007; 26: 4518-4524.
- Cepus, M. Borth, M. Seitz, Int. J. Clean Coal and Energy. 2016; 5: 13-22.
- Khotele, N.T. Kalyani, S.J. Dhoble, Luminescence. 2018; 33, 1415-1423.
- Liu, Y. Sun, M.H. Lu, X.M. Pan, Z.B. Wang, J. Adhes. Sci. Technol. 2021; 36: 567-585.
- V. Sakthivel, P. Sankudevan, P. Vennila, G. Venkatesh, S. Kaya, G. Serdaroğlu, J. Mol. Struct. 2021; 1233: 130097-130100.
- Aravindan, K. Sivaraj, C. Kamal, P. Vennila, G. Venkatesh, J. Mol. Struct. 2021; 1229: 129488-129523.
- H. Meng, P. Zhou, F. Song, Y.B. Wang, G.L. Liu, H. Li, CrystEngComm. 15: 2786-2791.
- Li, H. Yan, K. Liu, X.Q. Huang, M.J. Niu, J. Mol. Struct. 2019; 1195: 470-478.
- X. Liu, Y. Liu, T.Z. Yu, W.M. Su, Y.Y. Niu, et al. Chem. Front. 2018; 5: 2321-2333.
- Z. Yu, Z.Y. Zhu, Y.J. Bao, Y.L. Zhao, X.X. Liu, H. Zhang, Dyes. Pigments. 2017; 147: 260-269.
- A. Ramakrishnan, World. J. Virol. 2016; 5: 85-86.
- E. Mihajlovic-Lalic, J. Poljarevic, S. Grguric-Sipka, Inorg.Chim. Acta. 2021; 527: 120582-120600.
- Umar, H. Yin, M. Luo, Ann. Clin. Med. Case. Rep. 2022; 10: 1-10.

