Research Article - Volume 3 - Issue 6
Analysis of erg 11 expression in clinical isolates of dermatophytes in pat ients with resistant tinea infection
Shyama Datt*
Department of Microbiology, University College of Medical Sciences & Guru Teg Bahadur Hospital, Delhi, India.
Received Date : Oct 05, 2023
Accepted Date : Oct 30, 2023
Published Date: Nov 06, 2023
Copyright:© Shyama Datt 2023
*Corresponding Author : Shyama Datt, Department of Microbiology, University College of Medical Sciences & Guru Teg Bahadur Hospital, Delhi, India.
Email: shyamadutt0@gmail.com
DOI: Doi.org/10.55920/2771-019X/1578
Abstract
Background and objectives: Dermatophytes are keratinophilic groups of microorganisms which invade keratinized tissue. Our objectives of the study are as follows.
Methods: Conventional phenotypic and genotypic methods were employed to identify the pathogenic dermatophyte isolate. Expression of Ergosterol 11 gene in culture isolates was done by Quantitative Real-Time PCR (qRT-PCR).
Results: A total of 120 dermatophytosis cases attending a dermatological clinic in a tertiary care hospital were included in the study. Microscopy of KOH mount was positive in 90 isolates while 60 were culture positive. The most common dermatophytes implicated were Trichophyton mentagrophytes (75%) followed by Trichophyton rubrum (25%) which were confirmed by PCR using the species-specific primer. Higher MIC was detected for fluconazole (66.67 %), itraconazole (6.67%), terbinafine (20%) and griseofulvin (10%), for T. mentagrophytes complex , while all strains showed lower MIC for voriconazole and luliconazole.
Interpretation & Conclusion: The study observed a predominance of T. mentagrophytes complex causing chronic dermatophytosis. The rising MIC to fluconazole and terbinafine among the isolates raises a concern for judicious use of antifungals for effective management. The real-time PCR analysis of ERG 11 expression in certain isolates demonstrated up-regulation in patients not responding to treatment as compared to ATCC T. mentagrophytes strains.
Abbreviations: MIC-minimal inhibitory concentration, MIC50- the lowest concentration of the antifungal at 50% of the isolates were inhibited, GM- Geometric mean.
Introduction
Dermatophytes, a group of keratinophilic filamentous fungi thriving on the keratin substrate, are the etiological agents responsible for causing superficial fungal infection in human and animals with an estimated global prevalence of approximately 20 percent as per the World Health Organization (WHO) report. Predominant in the tropical and subtropical countries; especially in the developing countries like India, the hot and humid climate is favorable to the acquisition and maintenance of the disease [1]. Routine procedures for dermatophyte species identification is based on the conventional phenotypic method of microscopy and culture. Morphological and physiological characteristics can frequently vary, often influenced by temperature variation, and type of medium, further hampering strain identification. In the last few years, genotypic approaches have proven to be useful for addressing taxonomic complexities in dermatophytes; moreover, genotypic detection is considered more stable and precise than phenotypic characteristics [2-5]. In the past few years, chronic dermatophytosis has become a public health problem. Recalcitrant dermatophytosis encompasses relapse, recurrence or re-infection & persistence of infection [6]. Systemic antifungals are used for treating dermatophytosis as monotherapy or in combination, and azoles are still used as the first line of management in several cases. Azoles inhibit the fungal cytochrome P-450 enzyme lanosterol 14-α-demethylase (Cyp51) encoded by ERG 11 gene, being effective against several fungi including the common dermatophytes like Trichophyton rubrum, Trichophyton mentagrophytes complex, and Epidermophyton floccosum as documented by Garvey et al. [7] Studies have reported increasing minimal inhibitory concentration (MIC) values to fluconazole, terbinafine and griseofulvin amongst dermatophytes leading to clinical failure. Excessive use of topical steroid, antifungal, antibacterial ointments available at pharmacy has aggravated the severity and further delayed healing. Moreover vellus hair involvement even if the body surface involvement is less, is an indicator of chronicity and hence requires systemic antifungal therapy. Further triazole resistant environmental hotspots can be possible sources for selection and reservoirs of azole-resistant fungal agents may facilitate their selection as proven as in A. fumigatus [8]. The potential loss of efficacy of azoles has prompted many researchers to make concerted efforts to discover new drugs that might block fungal growth at different metabolic sites [9,10]. Hence this study was aimed to evaluate the expression of ERG 11 genes by real-time PCR for fluconazole and itraconazole in dermatophytes isolated from patients previously exposed to antifungals and having persistent lesions residing in East Delhi compared to the laboratory maintained T. mentagrophytes ATCC 28185 strain.
Materials and Methods
The present study was conducted on a total of 120 samples from clinically diagnosed dermatophytosis patients, subjected to culture from skin and nail samples of patients attending dermatology OPD of a tertiary care hospital, Delhi from January 2016 to December 2018. A portion of each clinical specimen was suspended in a drop of 10% & 40% potassium hydroxide (KOH) for processing skin and nail respectively. KOH wet mount slides were viewed under a light microscope under 40X magnification. A portion of the sample was cultured on Sabouraud’s dextrose agar (Hi-media, Mumbai) with antibiotics with chloramphenicol (0.05 g/l), gentamicin (20 mg/l) and cycloheximide (0.5 g/l). All inoculated tubes were incubated at 25°C for optimal growth. After growth, the etiological agent was confirmed by the characteristic morphology of the colony and by studying the microscopic appearance of the fungus on Lacto Phenol Cotton Blue (LPCB) mount and Urease test. T. mentagrophytes ATCC 28185 strain was used as control. The molecular confirmation of isolates was done by using speciesspecific primers of T. mentagrophytes and T. rubrum [11]. DNA extraction and PCR: DNA was extracted from the cultures grown on SDA by using the commercially available DNA extraction kit (HiYield Genomic DNA Kit, RBC, Taiwan). On the basis of alignment of sequences of internal transcribed spacer region ITS 1and 2 in the NCBI nucleotide database, a primer of T. rubrum forward (203bp) GACCGACGTTCCATCAGGGGT and reverse TCAGACTGACAGCTCTTCAGAG and T. mentagrophytes (130 bp) forward CAAACGTCCGTCAGGGTGAGC and reverse TAGCCACTAAAGAGAGGCTCGC were used for amplification [12,13]. Each tube contained a total volume of 25 μl which included 2.5 μl buffer (10X), 5 μl of Q-buffer, 0.5 μl dNTPs (200 μM), MgCl2 0.5 μl (1.5 mM), 0.15 μl Taq polymerase, 1 μl of each primer, forward and reverse (10 μM), 5 μl of the extracted DNA and nuclease-free water to make up the volume. All PCR reagents were from the Taq PCR Core Kit (Fisher Scientific-Qiagen, Germany). Amplification was performed in a Mastercycler personal (Eppendorf, Hamburg, Germany). Initial denaturation was performed at 94°C for 10 min which was followed by 35 amplification cycles of 30 s at 95°C and 45 s at 65°C and 30 s at 72°C, and final extension of 10 min at 72°C. The amplified PCR products were analyzed by electrophoresis on 1.5% agarose gel, stained with Ethidium bromide at 125 V and 15 mAh current in a 10 slot apparatus for 30 min. A molecular marker of 100 bp was used to determine the size of the amplicons [14,15]. Standard T. mentagrophytes ATCC 28185 control strains were used as positive controls [16].
RNA isolation: Total RNA from culture isolates obtained from skin and nail were extracted by TRIzol TM Reagent (Invitrogen Bio services India Pvt. Ltd) method. Isolates were mixed in TRIzol TM Reagent, 1 ml of RNA express reagent was added in the sample, total lysate cells were obtained, which was further mixed by micropipette to form a homogeneous lysate and incubated for 5 min at room temperature. 200µl of chloroform per ml of RNA express reagent was added and centrifuged at 12,000 rpm for 15 min at 4°C to obtain a colorless upper aqueous phase containing RNA which was separated in another 1.5 ml Eppendorf tube. RNA was washed by adding 500 µl of Isopropanol. The Eppendorf tube was centrifuged at 12,000 rpm for 10 min at 4°C and supernatant was discarded.1 ml of 75% ethanol was added to the pellet and vortexed. This was followed by centrifugation at 10,500 rpm for 5 min at 4°C. The supernatant was discarded, and the pellet was suspended in RNase free water, which was incubated at 55-60°C for 15 min. The isolated RNA was stored at -80°c immediately. The purity and integrity of the RNA samples were confirmed by agarose gel electrophoresis. Nano-drop determined the concentration and purity of the samples by measuring concentration and 260/280 ratio.
cDNA Preparation: -Extracted RNA was used as a template for cDNA synthesis by Superscript reverse transcriptase II (Invitrogen Bioservices India Pvt. Ltd) kit. Total RNA (2-5 µg) from each sample was reverse transcribed into cDNA.
Gene expression of Ergosterol 11 in culture isolates by Quantitative Real-Time PCR (qRT-PCR): To quantify expression of Ergosterol 11(ERG 11) and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), qRT-PCR was performed using the Light Cycler® 480 Instrument (Roche Diagnostics, Germany). Thirty isolates (Trichophyton mentagrophytes) were chosen from patients who were identified as clinically not responding to treatment , exposed to over the counter use of topical or systemic azole as antifungal drugs presenting to the OPD with extensive persisting lesions.
Real-Time PCR reaction mixtures contained: Light Cycler® 480 SYBR Green I Master Mix, Nuclease-free water (Roche Diagnostics, Germany), cDNA and primers. Primers are ERG 11 forward 5' CACTTCCTTGCCCTGTAGAGATC 3', ERG 11 Reverse 5' GGAGTTTTCAATGTCAGCAAGGTTT 3', GAPDH Forward 5'-ACGGCTTCGGTCGTATTGG 3' and GAPDH Reverse 5′-ATGTATTCGGCGATTTGGTCT 3'. The final volume of each reaction was 20µl; the reaction mixtures contained 2-5µl of cDNA, 20 µl of SYBER Green Universal Master Mix, and 160 nM of each primer, and the volume was brought to 20 µl with nuclease-free water. The Real-Time PCR condition had been standardized according to the melting temperature (Tm) of the primers. The Ergosterol-11 is the gene of interest and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which served as an endogenous control. The PCR program was 95°C for 10 min, followed by 40 cycles of 95°C for 10s and the annealing/extension at 60°C for 20s and 72°C for 10s. Following PCR, dissociation curve analysis was performed to verify that a single product was amplified. Real-time PCR was carried out in a Roche Light Cycler 480-II (Roche Diagnostics, Germany). Amplification Curve for each gene was shown, and the threshold cycle (Ct) value was calculated by the software. Following Real-time PCR, dissociation curve analysis was performed to verify that a single product was amplified [17].
Preparation Antifungal agents as per CLSI M-38 A2: Antifungal susceptibility testing was performed according to the Clinical Laboratory Standard Institute (CLSI) 38 A2 guidelines suggested for molds. T. mentagrophytes ATCC 28185 was used as control. RPMI 1640 medium and MOPS (4-Morpholine propane sulfonic acid) (Sigma Aldrich, USA), pH 7.0 was used as a medium for suspension of the isolates. The antifungal drugs tested were fluconazole, itraconazole, terbinafine, griseofulvin, luliconazole and voriconazole (Sigma Aldrich, USA). The initial inoculum suspension of the isolates (i.e., 1 - 3 x 106cells/ml) was prepared using the spectrophotometer to match the optical density of 80 % transmittance at 530 nm wavelength. The final concentration of the inoculum (1- 3 x 103 cells/ml) is prepared in 1: 50 dilution in RPMI. The test assay which was performed in 96 U bottom microtitre plate was incubated for 96 hours at 35°C in BOD incubator [18].
Endpoint determination: Endpoint determination values were performed visually every 24 hrs until the indication of growth in control drug-free well i.e. for 48 & 96 hrs. The lowest dilution of the drug, which inhibited the fungal growth, was taken as the MIC. For itraconazole drugs, MICs corresponded to the lowest drug concentration that resulted in a reduction in the growth of 80% inhibition in compared with their growth control. Terbinafine MICs corresponded to the lowest drug concentration that gave a reduction in growth of 100% and for Fluconazole, growth reduction up to 50 %. According to CLSI 38 A2, MIC50 was calculated by taking the drug concentration, where 50% inhibition of colony by drug at 48 hours [18]. Statistical analysis was done using Statistical Package for Social Sciences package (SPSS; Inc., Chicago, IL, USA; version 20.0). The analysis comprised of calculating means and proportions. The independent sample t-test was used to test the statistical significance of the data.
Study Design: Cross-Sectional Analytical Study.
Sample Size: 60 confirmed isolates of T. mentagrophytes & T. rubrum from cases of dermatophytosis.
The sample size of 60 has been found to be statistically appropriate for this study (sample size calculated using the standard formula Z2P(1-P)/d2 for a prevalence study, where Z = Z statistic for a level of confidence (the value of Z is set at 1.96), P = expected prevalence or proportion (in proportion of one; if 45%, P = 0.45), and d= precision (in proportion of one; if 125%, d = 0.125)).
Formula -
n = Z2 α/2 P (1-P)/d2
= 1.96*1.96*0.45*0.55/0.125*0.125
=60.8
P = 45% i.e. 0.45
1-P = 55% i.e. 0.55
d = 0.45*0.25/100=0.125
so, we considering P rate of T. mentagrophytes & T. rubrum in dermatophytosis confirmed cases is 45% with 25% relative error either the site with 95% confidence level is 60.8 i.e. total 60 cases is taken
Inclusion criteria: Patients with clinically suspected superficial fungal infection in skin and nail.
Exclusion criteria:
- Patients with any systemic disease or autoimmune disorder, immunodeficiency, malignant disease or any immunosuppressive treatment.
- Pregnant and lactating women.
Source: The present study was carried out in the Department of Microbiology and Dermatology UCMS and GTBH, New Delhi. Ethical clearance was obtained from the ethical committee of UCMS >B Hospital.
Results
A total of 120 samples were collected which included 25 from onychomycosis patients and 95 skin scrapings from Tinea corporis and cruris patients. The mean age of patients was 32.5±11.32 year (with the range from 16 to 60 years). The duration of dermatophytic infection ranged from 3 months to 10 years (9.8 months ± 5.45 months). Out of 120 samples, 90 were found to be KOH positive, of which 60 samples were culture positive. The clinical profile of patients with culture-positive isolates (60) varied from those with the first episode of dermatophytic infection [1], those with no clinical cure despite antifungal therapy (55) and some with relapse [4].
Of the 60 isolates, 15(25%) isolates were identified as Trichophyton rubrum and 45(75%) isolates as Trichophyton mentagrophytes complex on phenotypic mycological assessment. T. rubrum was the predominant pathogen (66.67%) isolated from nail samples than from skin samples of Tinea corporis & Tinea pedis (20 %). T. mentagrophytes complex was isolated in majority from T. corporis and T. cruris (44.44 %) followed by T. faciae (13.33 %), T. pedis (11.11 %) and onychomycosis (4.44 % ). All isolates were also subjected to conventional PCR using the species-specific primer of their ITS region for further confirmation, which showed 100% agreement as in Figure A & B.
Real-time PCR was performed for 30 isolates and assessed for ERG 11 expression as fold change (by calculating 2−ΔΔCT value) compared to T. mentagrophytes ATCC 28185 strain as control. The Ct value of ERG 11 gene was normalized by Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH). The fold change in the level of expression of ERG 11 genes using Real-Time PCR data and by applying 2−ΔΔCT method was 19.99-fold change and up-regulated between the clinical isolates and T. mentagrophytes ATCC 28185 strain. (Table I)
On antifungal susceptibility testing of these 30 isolates , the minimum inhibitory concentration (MIC) range for T. mentagrophytes complex showed a maximum of sensitivity to fluconazole <64 µg/ml (33.33%); itraconazole (93.33%) at <1 µg/ml and terbinafine (80%) at <1 µg/ml . All strains showed lower MIC for voriconazole and luliconazole as depicted in Table II. Higher MIC was detected for fluconazole (66.67 %), itraconazole (6.67 %), terbinafine (20 %) and griseofulvin (10 %), for T. mentagrophytes complex, while all strains showed 100% susceptibility for voriconazole and luliconazole . Most of the patients with the history of recurrence exhibited high MIC for either one or all the three drugs tested.
Discussion
Chronic dermatophytosis has emerged as a significant problem in India with several cases of reinfection and relapse and failure to the treatment being reported across the country. The present study was conducted in patients with dermatophytosis, attending the dermatology OPD of a tertiary care hospital in East Delhi, India. The geographical condition of this part of our country has provided a favorable environment for the survival of the dermatophytes and adaptation to the human host for several decades [19]. Besides the conducive environment, other factors that support the survival of the fungal agent, include unhygienic living condition, high population density, application of steroids, incomplete treatment and probable drug resistance [20]. As documented in other studies, males appeared to be more exposed to acquire dermatophytic infection (58.3 %) as compared to females (41.7%) which is in accordance with the other researchers worldwide [21]. Interestingly we found T. mentagrophytes complex was more prevalent than T. rubrum causing dermatophytosis in Delhi, similar to other studies [22]. The ongoing challenges in nomenclature with the different taxonomic approaches hinder the appropriate detection of species complicating the problem further for laboratories to make the correct identification [23-25]. As per de Hoog et al, the current taxonomy is Trichophyton mentagrophytes complex, which includes both Trichophyton mentagrophytes and Trichophyton interdigitale [23]. However Pietro Nenoff et al pointed out the mistaken identification of Trichophyton isolates is not limited to India but very likely to occur due to the lack of appropriate molecular diagnosis and lack of updated database [24, 25]. Though therapy for dermatophytosis is based on extent of lesion, severity, anatomical site and other underlying host factors for deciding upon the choice of antifungals by the clinicians, hence changes in taxonomy may not be a deciding factor for initiating management; rather it is important for epidemiology, geographical distribution and pathogenesis.
Table 1: Fold change in the level of expression of Erg 11 genes using Real-Time PCR data and by applying 2−ΔΔCT method.
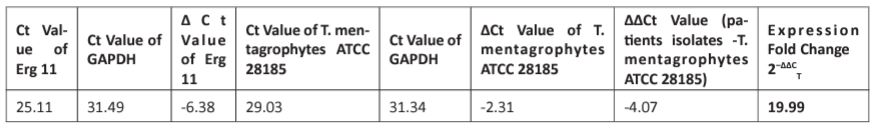
Table 2: In vitro susceptibility pattern of dermatophyte isolates (n = 30) against 4 antifungal drugs.

Out of 60 patients, 1.7% of cases were naive cases presenting with the first episode of dermatophytic infection, 91.7 % of cases were previously on antifungals for 1-2 yrs and 6.6% cases categorized as relapse. In our study ,the MIC range for T. mentagrophytes for different antifungals were performed within range of 0.125µg/ml-128 µg/ml for fluconazole; 0.0313 µg/ml-32 µg/ml for itraconazole, terbinafine and griseofulvin; 0.0625 µg/ml-16 µg/ml for voriconazole and 0.000125 µg/ml-1 µg/ml for luliconazole. Higher MIC was detected for fluconazole (66.67 %) for T. mentagrophytes complex, which was also observed in other studies. [26] To further investigate cellular responses to perturbation of ergosterol biosynthesis, we detected the expression of ERG 11 genes by real-time RT-PCR analysis. In our study, ERG 11 was found to be up-regulated 19.9 fold in the 30 isolates recovered from patients with recalcitrant dermatophytosis. The poor outcome with persistence of lesion and increased inflammation are a common presentation seen in majority of these patients hence the effect of upregulated Erg 11 expression may have been a consequence of inadequate sub MIC dose exposure to azoles, often used as topical applications or is it an adaptation of the dermatophytes to environmental exposure to fungal pesticides is still a speculation. Fungal resistance to azole reagents has been attributed to various genetic mutations in its target gene erg11 (cyp51), and/or the upregulation of efflux pump genes such as MDR1, CDR1, and CDR2. Transcription factors acting on effector genes is another important attribution to drug resistance(s) characterized in a number of clinical species. [27] Up-regulation of sterol metabolism gene, ERG 11, which has been demonstrated in previous reports with Candida albicans, probably emphasizes the global up-regulation of sterol metabolism genes in response to ergosterol biosynthesis inhibitors [28-30]. The argument for overexpression is that accumulation of an early substrate or toxic sterol by-products induces the expression of genes responsible for sterol metabolism. The rise in ERG 11 expression in our study suggest a survival strategy of dermatophytes and exposure to azoles may have led to the rising MIC to fluconazole.
Ergosterol is an important constituent of cellular membranes and plays a fundamental role in many biological processes [31]. In the biosynthesis of ergosterol with the exception of ERG 7, most of the responsive genes function downstream of ERG 11, suggesting that their induction is in response to ergosterol depletion. This regulatory manner of gene expression in the ergosterol pathway is similar to that previously reported for ketoconazole exposure of C. albicans [32]. Because overexpression of the enzyme encoded by ERG 11 is known to contribute to azole resistance in C. albicans strains [29, 31, 32]. hence similar consequences may be responsible for rising MIC to azoles amongst dermatophytes [33].
The potential loss of efficacy of azoles has prompted concerted efforts to discover new drugs like Amorolphine that might block fungal growth at different metabolic sites. However, which mechanism is responsible for the global up-regulation of sterol metabolism genes in response to ergosterol biosynthesis inhibitors remains unclear. Hence the differential expression of selected genes responsible for cell wall or membrane synthesis by RT-PCR can be a useful tool in associating the rising MIC with clinical failure in patients of dermatophytosis [34,35].
Conclusion
Trichophyton mentagrophytes complex is more prevalent than Trichophyton rubrum in the population of Delhi NCR. Trichophyton mentagrophytes complex is closely associated with T. corporis and T. cruris affecting all age groups, especially in males. Azole resistance as demonstrated by high MIC and upregulated ERG11 expression in patients not responding to treatment raises serious concern regarding excessive use of azoles and warrants judicious use in clinical settings.
In conclusion, our knowledge of understanding the complexity of recalcitrant dermatophytosis extensive work on the molecular biology of dermatophytes (T. mentagrophytes complex and T. rubrum) is still lacking, and it is difficult to indicate antifungal resistance to be entirely responsible for the current status of resistant Tinea. Our study gives preliminary data on ERG 11 gene upregulation with a distinct difference between the clinical isolates and the wild type T. mentagrophytes ATCC. 28185 strain, though additional mutational studies will reveal the mechanism and other potential elements like efflux pump involved in antifungal resistance. Limitation of our study: Limited sample size couldn’t reflect in correlation between ERG11 gene expression and MICs of isolates. Further studies with appropriate sample size are needed to delve into the nature of antifungal resistance in dermatophytosis.
Acknowledgment
This work was financially supported by the Indian Council of Medical Research, New Delhi (Project No. 80/970/2015-ECD-I).
The authors report no conflicts of interest.
References
- Marques SA, Robles AM, Tortorano AM, Tuculet MA, Negroni R, Mendes RP. Mycoses associated with AIDS in the Third World. Sabouraudia. 2000; 38 (1): 269-79.
- Weitzman, Irene; Summerbell, Richard C. The dermatophytes. Clinical microbiology reviews. 1995; 2: 240-259.
- Brillowska-Dąbrowska A, Saunte DM, Arendrup MC. Five-hour diagnosis of dermatophyte nail infections with specific detection of Trichophyton rubrum. Journal of clinical microbiology. 2007; 45 (4): 1200-4.
- Faggi E, Pini G, Campisi E. PCR fingerprinting for identification of common species of dermatophytes. Journal of clinical microbiology. 2002; 40(12): 4804-5.
- Liu D, Pearce L, Lilley G, Coloe S, Baird R, Pedersen J. PCR identification of dermatophyte fungi Trichophyton rubrum, T. soudanense and T. gourvilii. Journal of medical microbiology. 2002; 51(2): 117-22.
- Sardana K, Mahajan K, Arora P. Superficial fungal infections. In: Sardana K, Mahajan K, Arora P, editors. Fungal infection diagnosis and treatment. 1st edition. CBS Publishers & Distributors Pvt Ltd. 2017; 81-2.
- Garvey EP, Hoekstra WJ, Moore WR, Schotzinger RJ, Long L, Ghannoum MA. VT-1161 Dosed Once-Daily or Once-Weekly Exhibits Potent Efficacy in the Treatment of Dermatophytosis in a Guinea Pig Model. Antimicrobial agents and chemotherapy. 2015 ; AAC-04902.
- Schoustra SE, Debets AJ, Rijs AJ, Zhang J, Snelders E, Leendertse PC, et al. Environmental Hotspots for Azole Resistance Selection of Aspergillus fumigatus, the Netherlands. Emerging infectious diseases. 2019; 25(7).
- Yamada T, Maeda M, Alshahni MM, Tanaka R, Yaguchi T, Bontems O, et al. Terbinafine resistance of Trichophyton clinical isolates caused by specific point mutations in the squalene epoxidase gene. Antimicrobial agents and chemotherapy. 2017; AAC-00115.
- Hoekstra WJ, Garvey EP, Moore WR, Rafferty SW, Yates CM, Schotzinger RJ. Design and optimization of highly-selective fungal CYP51 inhibitors. Bioorganic & medicinal chemistry letters. 2014; 24(15): 3455-8.
- Singh S, Beena PM. Comparative study of different microscopic techniques and culture media for the isolation of dermatophytes. Indian journal of medical microbiology. 2003; 21(1): 21.
- Nagao K, Sugita T, Ouchi T, Nishikawa T. Identification of Trichophyton rubrum by nested PCR analysis from paraffin embedded specimen in trichophytia profunda acuta of the glabrous skin. Nippon Ishinkin Gakkai Zasshi. 2005; 46(2): 129-32.
- Gräser Y, El Fari M, Presber W, Sterry W, Tietz HJ. Identification of common dermatophytes (Trichophyton, Microsporum, Epidermophyton) using polymerase chain reactions. British Journal of Dermatology. 1998; 138 (4): 576-82.
- Faggi E, Pini G, Campisi E, Bertellini C, Difonzo E, Mancianti F. Application of PCR to distinguish common species of dermatophytes. Journal of clinical microbiology. 2001; 39(9): 3382-5.
- Kanbe T, Suzuki Y, Kamiya A, Mochizuki T, Fujihiro M, Kikuchi A. PCR-based identification of common dermatophyte species using primer sets specific for the DNA topoisomerase II genes. Journal of dermatological science. 2003; 32(2): 151-61.
- Ghannoum M, Chaturvedi V, Diekema D, Ostrosky-Zeichner L, Rennie R, Walsh T, et al. Multilaboratory evaluation of in vitro antifungal susceptibility testing of dermatophytes for ME1111. Journal of clinical microbiology. 2015: JCM-03019.
- Liang RM, Cao YB, Fan KH, Xu Y, Gao PH, Zhou YJ, et al. 2-Amino-nonyl-6-methoxyl-tetralin muriate inhibits sterol C-14 reductase in the ergosterol biosynthetic pathway. Acta Pharmacologica Sinica. 2009; 30(12): 1709.
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi; Approved Standard -Document M38-A2. 2nd ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2008.
- Khan S, Singhal S, Mathur T, Upadhyay DJ, Rattan A. Antifungal susceptibility testing method for resource constrained laboratories. Indian journal of medical microbiology. 2006 Jul 1; 24 (3): 171.
- Patel P, Mulla S, Patel D, Shrimali G. A study of superficial mycosis in south Gujarat region. Natl J Commun Med. 2010; 1 (2): 85-8.
- Tigga RA, Das S, Bhattacharya SN, Saha R, Pandhi D, Datt S, et al. Burden of Chronic Dermatophytosis in a Tertiary Care Hospital: Interaction of Fungal Virulence and Host Immunity. Mycopathologia. 2018; 183(6): 951-9.
- Dabas Y, Xess I, Singh G, Pandey M, Meena S. Molecular identification and antifungal susceptibility patterns of clinical dermatophytes following CLSI and EUCAST guidelines. Journal of Fungi. 2017; 3(2): 17.
- de Hoog GS, Dukik K, Monod M, Packeu A, Stubbe D, Hendrickx M, et al. Toward a novel multilocus phylogenetic taxonomy for the dermatophytes. Mycopathologia. 2017; 182(1-2): 5-31.
- Nenoff P, Verma SB, Uhrlaß S, Burmester A, Gra¨ser Y. A clarion call for preventing taxonomical errors of dermatophytes using the example of the novel Trichophyton mentagrophytes genotype VIII uniformly isolated in the Indian epidemic of superficial dermatophytosis. Mycoses. 2018. https://doi.org/10.1111/myc.12848.
- Chowdhary A, Singh A, Singh PK, Khurana A, Meis JF. Perspectives on misidentification of Trichophyton interdigitale/Trichophyton mentagrophytes using internal transcribed spacer region sequencing: Urgent need to update the sequence database. Mycoses. 2019; 62(1): 11-5.
- Sardana K, Kaur R, Arora P, Goyal R, Ghunawat S. Is antifungal resistance a cause for treatment failure in dermatophytosis: A study focused on tinea corporis and cruris from a tertiary centre?. Indian dermatology online journal. 2018; 9(2): 90.
- Liu J, Wang S, Qin T, Li N, Niu Y, Li D, et al. Whole transcriptome analysis of Penicillium digitatum strains treatmented with prochloraz reveals their drug-resistant mechanisms. BMC genomics. 2015; 16(1): 855.
- Mendes NS, Bitencourt TA, Sanches PR, Silva-Rocha R, Martinez-Rossi NM, Rossi A. Transcriptome-wide survey of gene expression changes and alternative splicing in Trichophyton rubrum in response to undecanoic acid. Scientific reports. 2018; 8(1): 2520.
- Khurana A, Masih A, Chowdhary A, Sardana K, Borker S, Gupta A, et al. Correlation of in vitro susceptibility based on MICs and SQLE mutations with clinical response to terbinafine in patients with tinea corporis/cruris. bioRxiv. 2018; 326603.
- Rudramurthy SM, Shankarnarayan SA, Dogra S, Shaw D, Mushtaq K, Paul RA, et al. Mutation in the Squalene epoxidase gene of Trichophyton interdigitale and Trichophyton rubrum associated with allylamine resistance. Antimicrobial agents and chemotherapy. 2018; AAC-02522.
- De Backer MD, Ilyina T, Ma XJ, Vandoninck S, Luyten WH, Bossche HV. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrobial agents and chemotherapy. 2001; 45(6): 1660-70.
- Perfect JR. The antifungal pipeline: a reality check. Nature reviews Drug discovery. 2017; 16(9): 603.
- Diao Y, Zhao R, Deng X, Leng W, Peng J, Jin Q. Transcriptional profiles of Trichophyton rubrum in response to itraconazole. Medical mycology. 2009; 47(3): 237-47.
- Salehi Z, Shams-Ghahfarokhi M, Razzaghi-Abyaneh M. Antifungal drug susceptibility profile of clinically important dermatophytes and determination of point mutations in terbinafine-resistant isolates. European Journal of Clinical Microbiology & Infectious Diseases. 2018; 37(10): 1841-6.
- Martinez-Rossi NM, Bitencourt TA, Peres NT, Lang EA, Gomes EV, Quaresemin NR, et al. Dermatophyte Resistance to Antifungal Drugs: Mechanisms and Prospectus. Frontiers in microbiology. 2018; 9: 1108.

