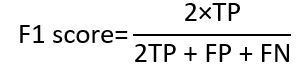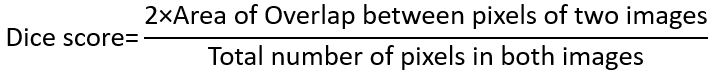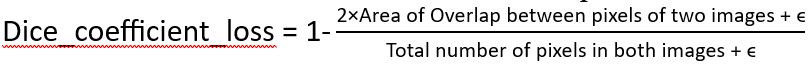Research article - Volume 4 - Issue 2
An application of deep learning-based method (U-net) for left atrial segmentation from magnetic resonance imaging (MRI)
Muhammad Adib Uz Zaman*
School of Information Technology, University of Cincinnati, USA.
Received Date : Jan 26, 2024
Accepted Date : Feb 26, 2024
Published Date: Mar 04, 2024
Copyright: © Zaman MAU 2024
*Corresponding Author : Muhammad Adib Uz Zaman* School of Information Technology, University of Cincinnati, USA.
Email: a_u_z_ipe@yahoo.com
DOI: Doi.org/10.55920/2771-019X/1640
Abstract
This study aims to provide significant insights into the automatic left atrium (LA) segmentation from MRI images of atrial fibrillation (AF) patients using U-net that is a deep learning-based image segmentation technique. This segmentation task is important because it will allow researchers to simulate LA activities in real life before, during or after AF episodes. The dataset used in this study contains the MRI images from patients who went through catheter ablation as part of their prognosis of AF and then a follow up period ranging from 3 to 7 months. U-net performed well for the LA segmentation tasks across different MRI slices during training and testing.
Introduction and background
Precise understanding of the anatomical structure of the LA can provide guidance for the catheter ablation technique and enhance the success rate of the surgery. Gadolinium-Enhanced Magnetic Resonance Imaging (GE-MRI) is a well-established and non-invasive method that provides clear pictures of the interior structure. It has a wide range of applications in examining the amount of fibrosis in the left atrium. The problem in segmenting the left atrium (LA) with GE-MRI is the limited contrast between the LA and the background, which is caused by the suppression of signal from healthy tissue. Most current studies on LA segmentation heavily depend on manual segmentation of GEMRI images [1]. The researchers have faced multiple challenges while segmenting LA from MRI slices [2]. Within the framework of the deep learning technique, sliding windows on appearance features are utilized to construct semantic information. In the domain of computer vision, it has met with a great degree of success and is capable of fitting practically any polynomial function [3, 4]. Recently, there has been a significant increase in the utilization of techniques that are founded on deep learning in the field of medical picture segmentation. On the other hand, the bulk of the applications involve the lungs, the brain, the left ventricle (LV), and other organs [5].
Data description
The MRI slices came from comprehensive arrhythmia research and management (CARMA), Center for Integrative Biomedical Computing (CIBC) and National Institute of health (NIH) [2, 6, 7]. Only a portion of the data was trained on U-net and evaluated using 16 volumes of 3D GE-MRI slices. The Received: Jan 26, 2024 Accepted: Feb 26, 2024 Published Online: Mar 04, 2024 Copyright: © Zaman MAU (2024). This Article is distributed under the terms of Creative Commons Attribution 4.0 Internat ional License. Cite this article: Zaman MAU. An application of deep learningbased method (U-net) for left atrial segmentation from magnetic resonance imaging (MRI). J Clin Med Images Case Rep. 2024; 4(1): 1640. dimensions of each volume vary depending on the subject, but all include at least 15-20 sequences in the Z-axis. We divided the 3D volumes into 2D slices along the Z axis and center cropped them to 338 × 338. Validation set comprised 10% of training slices. Ultimately, we obtained more than 347 resized MRI slices for the experiment.
Methods
The original U-Net model was developed specifically for the purpose of segmenting cells. The challenge in cell segmentation lies in the indistinct demarcation between the cells. Nevertheless, notwithstanding the issue of hazy boundaries, LA segmentation continues to face the challenge of imbalanced distribution between positive and negative samples. The background comprises most of the LA pictures. If this problem persists, the model will not learn the required useful characteristics. To address this issue of imbalance, there were some modifications to the original U-Net model by including the dice coefficient loss function [1]. In usual classification task (segmentation is somewhat like binary classification), the following metric called F1 score is heavily used:
But image segmentation benefits from the use of dice score as shown below:
The modified U-net method introduces a unique dice coefficient [1] as shown:
Here, ϵ is a positive number that boosts the convergence speed for U-net. As mentioned previously, each patient has more than 10 slices. These slices are cropped to the center and a 338 x 338 box was used for cropping. Originally, each patient had more than 40 slices per MRI. However, some of these slices did not contain any LA information and were practically blank. That is why those slices with no or very less information were discarded and were not fed to the U-net to improve the data balancing.
Results
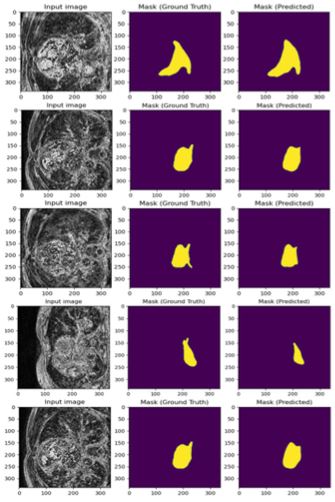
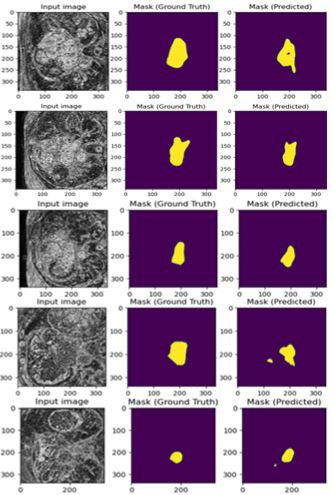
The results for the experiment are shown in Figure 1 and 2. As seen in the figures, the input image is one slice of MRI from one patient. The task was to detect the binary masks for that input image through U-net. Due to space limitation, all sample results were not included in this article. Figure 1 shows the training result and Figure 6 shows the corresponding testing result. Both results are quite close, and this verifies that there was no overfitting of the data. A total 74 epochs were run with a considerably small step length of 10^(-6) . It took around 3-4 hours to complete the training and produce the results.Figure 5 shows how close the training results were compared to the ground truth (input mask). Figure 2, however, does not show superior performance but given the nature of the data and low number of data-rich volumes, the results shown here are still quite good. As always, there is still room for improvement in this experimental study.
Figure 1 : Input image (338 × 338), ground truth and predicted mask (Training).
Figure 2 : Input image (338 × 338), ground truth and predicted mask (Testing).
Conclusion
LA segmentation is a challenging task that requires undivided attention and efforts from researchers in this domain. Artificial intelligence (AI) and deep learning methods can revolutionize this field as seen from the examples. However, it is also important to think about the ethical use of AI or deep learning methods. The success of AI hugely depends on the quality of the training data which is very difficult to collect. There is also a need for filtering out noisy data so that those did not impact the training and efficiency of deep learning models like U-net. In this experiment, the dice score is improved but still not sufficient for real life implementation. In future, the research direction should focus on the improvement of training data and prediction by ensuring higher dice scores for LA segmentation task
References
- Y Liu, Y Dai, C. Yan and K. Wang. Deep Learning Based Method for Left Atrial Segmentation in GE-MRI. Lecture Notes in Computer Science. 2019; 311-318.
- M Zaman and D. Du. A Review on Atrial Fibrillation (Computer Simulation and Clinical Perspectives). Hearts. 2022; 3: 20-37.
- A. A. A. C. F. L. G. G. P. J. C. V. R. S. J. G. B. Setio. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE transactions on medical imaging. 2016; 35: 1160-1169.
- Z Xiong, Q Xia, Z Hu, N Huang, C Bian, Y Zheng and J Zhao, “A global benchmark of algorithms for segmenting the left atrium from late gadolinium-enhanced cardiac magnetic resonance imaging,” Medical Image Analysis. 2021; 67: 101832.
- X. W. Y. S. G. L. Z. Z. Y. F. Y. Zhao.A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Medical image analysis. 2019; 43: 98-111.
- RHRJ Karim, M Balasubramaniam, Z Chen, D Perry, A Uddin and K Rhode. Evaluation of current algorithms for segmentation of scar tissue from late gadolinium enhancement cardiovascular magnetic resonance of the left atrium: an open-access grand challenge. Journal of Cardiovascular Magnetic Resonance. 2013; 15: 1-17.
- C McGann, N Akoum, A Patel, E Kholmovski, P Revelo, K Damal and N F Marrouche. Atrial fibrillation ablation outcome is predicted by left atrial remodeling on MRI,” Circulation: Arrhythmia and Electrophysiology. 2014; 7: 23-30.